Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
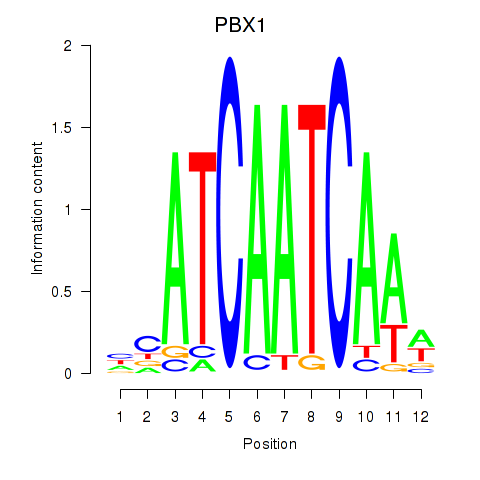
Results for PBX1
Z-value: 0.85
Transcription factors associated with PBX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PBX1
|
ENSG00000185630.14 | PBX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PBX1 | hg19_v2_chr1_+_164529004_164529060 | -0.13 | 5.9e-02 | Click! |
Activity profile of PBX1 motif
Sorted Z-values of PBX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_140135665 | 23.24 |
ENST00000340384.4
|
TUBB4B
|
tubulin, beta 4B class IVb |
| chr4_+_83956237 | 14.04 |
ENST00000264389.2
|
COPS4
|
COP9 signalosome subunit 4 |
| chr4_+_83956312 | 14.00 |
ENST00000509317.1
ENST00000503682.1 ENST00000511653.1 |
COPS4
|
COP9 signalosome subunit 4 |
| chr18_+_29171689 | 12.57 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr8_+_98788003 | 11.57 |
ENST00000521545.2
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr10_-_101190202 | 8.94 |
ENST00000543866.1
ENST00000370508.5 |
GOT1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr3_+_159557637 | 8.26 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr4_+_69962185 | 8.00 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr16_+_15596123 | 7.99 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr4_+_69962212 | 7.66 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr12_+_20968608 | 7.22 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr12_+_79258444 | 7.06 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr2_-_200323414 | 6.90 |
ENST00000443023.1
|
SATB2
|
SATB homeobox 2 |
| chr17_+_8191815 | 6.85 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr2_-_200322723 | 6.44 |
ENST00000417098.1
|
SATB2
|
SATB homeobox 2 |
| chr10_+_60028818 | 6.41 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr4_-_72649763 | 6.32 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chr14_+_50234309 | 6.28 |
ENST00000298307.5
|
KLHDC2
|
kelch domain containing 2 |
| chr2_-_10587897 | 6.21 |
ENST00000405333.1
ENST00000443218.1 |
ODC1
|
ornithine decarboxylase 1 |
| chr6_+_151561085 | 6.15 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chrX_-_92928557 | 6.11 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr8_-_91095099 | 6.01 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr6_+_151561506 | 5.96 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr15_-_37393406 | 5.79 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr2_+_181845298 | 5.61 |
ENST00000410062.4
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chrX_+_119737806 | 5.55 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr3_+_159570722 | 5.48 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr12_+_79258547 | 5.37 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr5_-_86708670 | 5.18 |
ENST00000504878.1
|
CCNH
|
cyclin H |
| chr3_+_139063372 | 5.14 |
ENST00000478464.1
|
MRPS22
|
mitochondrial ribosomal protein S22 |
| chr5_-_86708833 | 5.12 |
ENST00000256897.4
|
CCNH
|
cyclin H |
| chr22_+_31795509 | 5.12 |
ENST00000331457.4
|
DRG1
|
developmentally regulated GTP binding protein 1 |
| chr12_+_21168630 | 5.03 |
ENST00000421593.2
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr7_-_7680601 | 4.96 |
ENST00000396682.2
|
RPA3
|
replication protein A3, 14kDa |
| chr9_-_32573130 | 4.86 |
ENST00000350021.2
ENST00000379847.3 |
NDUFB6
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6, 17kDa |
| chr9_-_101471479 | 4.72 |
ENST00000259455.2
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr6_-_27114577 | 4.68 |
ENST00000356950.1
ENST00000396891.4 |
HIST1H2BK
|
histone cluster 1, H2bk |
| chrM_+_4431 | 4.62 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr19_+_33072373 | 4.58 |
ENST00000586035.1
|
PDCD5
|
programmed cell death 5 |
| chr2_-_96971232 | 4.55 |
ENST00000323853.5
|
SNRNP200
|
small nuclear ribonucleoprotein 200kDa (U5) |
| chr1_-_28241024 | 4.49 |
ENST00000313433.7
ENST00000444045.1 |
RPA2
|
replication protein A2, 32kDa |
| chr11_+_34938119 | 4.40 |
ENST00000227868.4
ENST00000430469.2 ENST00000533262.1 |
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr2_+_27886330 | 4.38 |
ENST00000326019.6
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr10_+_48189612 | 4.32 |
ENST00000453919.1
|
AGAP9
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9 |
| chr12_-_54653313 | 4.32 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr1_-_28241226 | 4.30 |
ENST00000373912.3
ENST00000373909.3 |
RPA2
|
replication protein A2, 32kDa |
| chr8_-_101965559 | 4.09 |
ENST00000353245.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr1_-_3713042 | 4.08 |
ENST00000378251.1
|
LRRC47
|
leucine rich repeat containing 47 |
| chr2_-_88427568 | 4.04 |
ENST00000393750.3
ENST00000295834.3 |
FABP1
|
fatty acid binding protein 1, liver |
| chr4_+_69681710 | 3.83 |
ENST00000265403.7
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr2_-_190044480 | 3.73 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr20_+_44441271 | 3.68 |
ENST00000335046.3
ENST00000243893.6 |
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr17_-_26694979 | 3.65 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr17_-_26695013 | 3.63 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr13_-_46679185 | 3.62 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr13_-_46679144 | 3.51 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr10_+_114709999 | 3.42 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr15_-_64673630 | 3.38 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr21_-_35014027 | 3.35 |
ENST00000399442.1
ENST00000413017.2 ENST00000445393.1 ENST00000417979.1 ENST00000426935.1 ENST00000381540.3 ENST00000361534.2 ENST00000381554.3 |
CRYZL1
|
crystallin, zeta (quinone reductase)-like 1 |
| chrX_-_140673133 | 3.23 |
ENST00000370519.3
|
SPANXA1
|
sperm protein associated with the nucleus, X-linked, family member A1 |
| chr16_-_49890016 | 3.22 |
ENST00000563137.2
|
ZNF423
|
zinc finger protein 423 |
| chrX_+_56590002 | 3.18 |
ENST00000338222.5
|
UBQLN2
|
ubiquilin 2 |
| chr5_+_162864575 | 3.16 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr19_+_14640372 | 3.13 |
ENST00000215567.5
ENST00000598298.1 ENST00000596073.1 ENST00000600083.1 ENST00000436007.2 |
TECR
|
trans-2,3-enoyl-CoA reductase |
| chr12_-_102455846 | 3.10 |
ENST00000545679.1
|
CCDC53
|
coiled-coil domain containing 53 |
| chr19_-_49955050 | 3.03 |
ENST00000262265.5
|
PIH1D1
|
PIH1 domain containing 1 |
| chr10_+_95517660 | 3.03 |
ENST00000371413.3
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chr17_+_72983674 | 2.99 |
ENST00000337231.5
|
CDR2L
|
cerebellar degeneration-related protein 2-like |
| chr14_-_74551096 | 2.95 |
ENST00000350259.4
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr8_+_37887772 | 2.95 |
ENST00000338825.4
|
EIF4EBP1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr2_-_27886460 | 2.94 |
ENST00000404798.2
ENST00000405491.1 ENST00000464789.2 ENST00000406540.1 |
SUPT7L
|
suppressor of Ty 7 (S. cerevisiae)-like |
| chr10_+_95517566 | 2.93 |
ENST00000542308.1
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chr10_+_5488564 | 2.87 |
ENST00000449083.1
ENST00000380359.3 |
NET1
|
neuroepithelial cell transforming 1 |
| chr16_-_70323422 | 2.83 |
ENST00000261772.8
|
AARS
|
alanyl-tRNA synthetase |
| chr2_+_219110149 | 2.81 |
ENST00000456575.1
|
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr2_+_79412357 | 2.78 |
ENST00000466387.1
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2 |
| chr11_-_14380664 | 2.78 |
ENST00000545643.1
ENST00000256196.4 |
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr19_+_41770236 | 2.71 |
ENST00000392006.3
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr4_-_70080449 | 2.71 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr9_+_106856831 | 2.70 |
ENST00000303219.8
ENST00000374787.3 |
SMC2
|
structural maintenance of chromosomes 2 |
| chr6_+_26199737 | 2.68 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chr3_+_179280668 | 2.54 |
ENST00000429709.2
ENST00000450518.2 ENST00000392662.1 ENST00000490364.1 |
ACTL6A
|
actin-like 6A |
| chr10_+_95517616 | 2.51 |
ENST00000371418.4
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chr12_-_102455902 | 2.46 |
ENST00000240079.6
|
CCDC53
|
coiled-coil domain containing 53 |
| chrX_-_70473957 | 2.42 |
ENST00000373984.3
ENST00000314425.5 ENST00000373982.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr1_-_54405773 | 2.40 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr6_-_26124138 | 2.38 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chrX_-_63005405 | 2.35 |
ENST00000374878.1
ENST00000437457.2 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr12_-_76478686 | 2.35 |
ENST00000261182.8
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr3_-_187388173 | 2.33 |
ENST00000287641.3
|
SST
|
somatostatin |
| chr2_-_242626127 | 2.31 |
ENST00000445261.1
|
DTYMK
|
deoxythymidylate kinase (thymidylate kinase) |
| chr12_+_12870055 | 2.30 |
ENST00000228872.4
|
CDKN1B
|
cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
| chrX_-_70474499 | 2.29 |
ENST00000353904.2
|
ZMYM3
|
zinc finger, MYM-type 3 |
| chr2_+_264869 | 2.26 |
ENST00000272067.6
ENST00000272065.5 ENST00000407983.3 |
ACP1
|
acid phosphatase 1, soluble |
| chr4_-_140005443 | 2.25 |
ENST00000510408.1
ENST00000420916.2 ENST00000358635.3 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr11_-_118966167 | 2.23 |
ENST00000530167.1
|
H2AFX
|
H2A histone family, member X |
| chr16_+_69458537 | 2.19 |
ENST00000515314.1
ENST00000561792.1 ENST00000568237.1 |
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane) |
| chr11_+_44117260 | 2.10 |
ENST00000358681.4
|
EXT2
|
exostosin glycosyltransferase 2 |
| chr19_-_16008880 | 2.10 |
ENST00000011989.7
ENST00000221700.6 |
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chrX_+_140677562 | 2.05 |
ENST00000370518.3
|
SPANXA2
|
SPANX family, member A2 |
| chr2_+_211342432 | 2.05 |
ENST00000430249.2
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr4_-_140005341 | 2.04 |
ENST00000379549.2
ENST00000512627.1 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr11_-_34938039 | 2.01 |
ENST00000395787.3
|
APIP
|
APAF1 interacting protein |
| chr3_+_129159039 | 2.01 |
ENST00000507564.1
ENST00000431818.2 ENST00000504021.1 ENST00000349441.2 ENST00000348417.2 ENST00000440957.2 |
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr5_-_10761206 | 2.00 |
ENST00000432074.2
ENST00000230895.6 |
DAP
|
death-associated protein |
| chr19_-_39881777 | 1.98 |
ENST00000595564.1
ENST00000221265.3 |
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr19_+_41770269 | 1.97 |
ENST00000378215.4
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr16_+_16484691 | 1.91 |
ENST00000344087.4
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr6_+_26124373 | 1.88 |
ENST00000377791.2
ENST00000602637.1 |
HIST1H2AC
|
histone cluster 1, H2ac |
| chr14_-_60097297 | 1.87 |
ENST00000395090.1
|
RTN1
|
reticulon 1 |
| chr19_-_49121054 | 1.84 |
ENST00000546623.1
ENST00000084795.5 |
RPL18
|
ribosomal protein L18 |
| chr2_+_264913 | 1.84 |
ENST00000439645.2
ENST00000405233.1 |
ACP1
|
acid phosphatase 1, soluble |
| chr22_-_39190116 | 1.79 |
ENST00000406622.1
ENST00000216068.4 ENST00000406199.3 |
SUN2
DNAL4
|
Sad1 and UNC84 domain containing 2 dynein, axonemal, light chain 4 |
| chr5_+_151151471 | 1.77 |
ENST00000394123.3
ENST00000543466.1 |
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr11_+_44117099 | 1.75 |
ENST00000533608.1
|
EXT2
|
exostosin glycosyltransferase 2 |
| chr1_+_205473720 | 1.69 |
ENST00000429964.2
ENST00000506784.1 ENST00000360066.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr22_-_30642782 | 1.69 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr10_+_124320195 | 1.68 |
ENST00000359586.6
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr10_+_18689637 | 1.67 |
ENST00000377315.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr3_+_129158926 | 1.65 |
ENST00000347300.2
ENST00000296266.3 |
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr11_+_844406 | 1.61 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr2_+_234600253 | 1.61 |
ENST00000373424.1
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr8_-_101965146 | 1.54 |
ENST00000395957.2
ENST00000395948.2 ENST00000457309.1 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr17_+_21030260 | 1.51 |
ENST00000579303.1
|
DHRS7B
|
dehydrogenase/reductase (SDR family) member 7B |
| chr2_+_12857015 | 1.41 |
ENST00000155926.4
|
TRIB2
|
tribbles pseudokinase 2 |
| chr14_+_29236269 | 1.39 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr5_-_43313574 | 1.39 |
ENST00000325110.6
ENST00000433297.2 |
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr6_+_27114861 | 1.38 |
ENST00000377459.1
|
HIST1H2AH
|
histone cluster 1, H2ah |
| chr5_+_65440032 | 1.35 |
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr14_+_104182061 | 1.30 |
ENST00000216602.6
|
ZFYVE21
|
zinc finger, FYVE domain containing 21 |
| chr5_+_140213815 | 1.30 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chrX_+_120181457 | 1.30 |
ENST00000328078.1
|
GLUD2
|
glutamate dehydrogenase 2 |
| chr11_+_71903169 | 1.29 |
ENST00000393676.3
|
FOLR1
|
folate receptor 1 (adult) |
| chr5_-_125930929 | 1.29 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr14_+_104182105 | 1.22 |
ENST00000311141.2
|
ZFYVE21
|
zinc finger, FYVE domain containing 21 |
| chr5_+_43602750 | 1.17 |
ENST00000505678.2
ENST00000512422.1 ENST00000264663.5 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr2_+_44396000 | 1.14 |
ENST00000409895.4
ENST00000409432.3 ENST00000282412.4 ENST00000378551.2 ENST00000345249.4 |
PPM1B
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
| chr7_-_72971934 | 1.13 |
ENST00000411832.1
|
BCL7B
|
B-cell CLL/lymphoma 7B |
| chr3_+_122785895 | 1.12 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5 |
| chr5_-_42825983 | 1.10 |
ENST00000506577.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr9_-_6015607 | 1.09 |
ENST00000259569.5
|
RANBP6
|
RAN binding protein 6 |
| chr6_-_167040731 | 1.08 |
ENST00000265678.4
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr4_+_37962018 | 1.05 |
ENST00000504686.1
|
PTTG2
|
pituitary tumor-transforming 2 |
| chr22_-_30866564 | 0.98 |
ENST00000435069.1
ENST00000415957.2 ENST00000540910.1 |
SEC14L3
|
SEC14-like 3 (S. cerevisiae) |
| chr17_+_73663402 | 0.97 |
ENST00000355423.3
|
SAP30BP
|
SAP30 binding protein |
| chr8_-_95487331 | 0.97 |
ENST00000336148.5
|
RAD54B
|
RAD54 homolog B (S. cerevisiae) |
| chr11_+_31531291 | 0.93 |
ENST00000350638.5
ENST00000379163.5 ENST00000395934.2 |
ELP4
|
elongator acetyltransferase complex subunit 4 |
| chr11_+_66384053 | 0.89 |
ENST00000310137.4
ENST00000393979.3 ENST00000409372.1 ENST00000443702.1 ENST00000409738.4 ENST00000514361.3 ENST00000503028.2 ENST00000412278.2 ENST00000500635.2 |
RBM14
RBM4
RBM14-RBM4
|
RNA binding motif protein 14 RNA binding motif protein 4 RBM14-RBM4 readthrough |
| chr10_+_124320156 | 0.88 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chrX_-_6453159 | 0.86 |
ENST00000381089.3
ENST00000398729.1 |
VCX3A
|
variable charge, X-linked 3A |
| chr11_+_22689648 | 0.85 |
ENST00000278187.3
|
GAS2
|
growth arrest-specific 2 |
| chr17_+_45973516 | 0.84 |
ENST00000376741.4
|
SP2
|
Sp2 transcription factor |
| chr5_-_132073210 | 0.78 |
ENST00000378735.1
ENST00000378746.4 |
KIF3A
|
kinesin family member 3A |
| chr16_-_20364030 | 0.75 |
ENST00000396134.2
ENST00000573567.1 ENST00000570757.1 ENST00000424589.1 ENST00000302509.4 ENST00000571174.1 ENST00000576688.1 |
UMOD
|
uromodulin |
| chr16_+_19222479 | 0.75 |
ENST00000568433.1
|
SYT17
|
synaptotagmin XVII |
| chr3_-_160283348 | 0.74 |
ENST00000334256.4
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3) |
| chr14_-_36789783 | 0.73 |
ENST00000605579.1
ENST00000604336.1 ENST00000359527.7 ENST00000603139.1 ENST00000318473.7 |
MBIP
|
MAP3K12 binding inhibitory protein 1 |
| chr5_+_151151504 | 0.68 |
ENST00000356245.3
ENST00000507878.2 |
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr6_+_167704838 | 0.68 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr4_-_69536346 | 0.67 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr17_+_73663470 | 0.66 |
ENST00000583536.1
|
SAP30BP
|
SAP30 binding protein |
| chr4_-_73434498 | 0.65 |
ENST00000286657.4
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr14_-_74551172 | 0.63 |
ENST00000553458.1
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr5_+_159656437 | 0.62 |
ENST00000402432.3
|
FABP6
|
fatty acid binding protein 6, ileal |
| chrX_+_140084756 | 0.61 |
ENST00000449283.1
|
SPANXB2
|
SPANX family, member B2 |
| chr15_-_64673665 | 0.59 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr12_-_57472522 | 0.56 |
ENST00000379391.3
ENST00000300128.4 |
TMEM194A
|
transmembrane protein 194A |
| chr16_-_29499154 | 0.54 |
ENST00000354563.5
|
RP11-231C14.4
|
Uncharacterized protein |
| chrX_-_110655306 | 0.54 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr1_+_117602925 | 0.52 |
ENST00000369466.4
|
TTF2
|
transcription termination factor, RNA polymerase II |
| chr3_+_148508845 | 0.50 |
ENST00000491148.1
|
CPB1
|
carboxypeptidase B1 (tissue) |
| chr2_-_154335300 | 0.50 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr5_-_132073111 | 0.49 |
ENST00000403231.1
|
KIF3A
|
kinesin family member 3A |
| chrX_-_102757802 | 0.48 |
ENST00000372633.1
|
RAB40A
|
RAB40A, member RAS oncogene family |
| chr10_+_5238793 | 0.47 |
ENST00000263126.1
|
AKR1C4
|
aldo-keto reductase family 1, member C4 |
| chr1_+_156589051 | 0.47 |
ENST00000255039.1
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2 |
| chr19_-_3978083 | 0.47 |
ENST00000600794.1
|
EEF2
|
eukaryotic translation elongation factor 2 |
| chr15_+_44069276 | 0.47 |
ENST00000381359.1
|
SERF2
|
small EDRK-rich factor 2 |
| chr13_-_33760216 | 0.45 |
ENST00000255486.4
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr12_+_21654714 | 0.45 |
ENST00000542038.1
ENST00000540141.1 ENST00000229314.5 |
GOLT1B
|
golgi transport 1B |
| chr3_-_169587621 | 0.43 |
ENST00000523069.1
ENST00000316428.5 ENST00000264676.5 |
LRRC31
|
leucine rich repeat containing 31 |
| chr16_-_20364122 | 0.39 |
ENST00000396138.4
ENST00000577168.1 |
UMOD
|
uromodulin |
| chr19_-_14640005 | 0.39 |
ENST00000596853.1
ENST00000596075.1 ENST00000595992.1 ENST00000396969.4 ENST00000601533.1 ENST00000598692.1 |
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr6_-_31782813 | 0.39 |
ENST00000375654.4
|
HSPA1L
|
heat shock 70kDa protein 1-like |
| chr7_+_134331550 | 0.38 |
ENST00000344924.3
ENST00000418040.1 ENST00000393132.2 |
BPGM
|
2,3-bisphosphoglycerate mutase |
| chr7_-_120498357 | 0.38 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr14_-_107211459 | 0.35 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chr18_+_580367 | 0.32 |
ENST00000327228.3
|
CETN1
|
centrin, EF-hand protein, 1 |
| chr5_+_43603229 | 0.30 |
ENST00000344920.4
ENST00000512996.2 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr11_-_34937858 | 0.27 |
ENST00000278359.5
|
APIP
|
APAF1 interacting protein |
| chr8_-_116681221 | 0.27 |
ENST00000395715.3
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr12_+_123944070 | 0.26 |
ENST00000412157.2
|
SNRNP35
|
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr19_-_39881669 | 0.24 |
ENST00000221266.7
|
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr3_+_193853927 | 0.24 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr4_+_120056939 | 0.23 |
ENST00000307128.5
|
MYOZ2
|
myozenin 2 |
| chr22_-_26961328 | 0.22 |
ENST00000398110.2
|
TPST2
|
tyrosylprotein sulfotransferase 2 |
| chr3_-_33481835 | 0.19 |
ENST00000283629.3
|
UBP1
|
upstream binding protein 1 (LBP-1a) |
| chr19_+_58341656 | 0.17 |
ENST00000442832.4
ENST00000594901.1 |
ZNF587B
|
zinc finger protein 587B |
| chr13_-_24895566 | 0.17 |
ENST00000422229.2
|
AL359736.1
|
protein PCOTH isoform 1 |
| chr4_-_105416039 | 0.14 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PBX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.4 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 3.0 | 8.9 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 1.9 | 5.6 | GO:0002188 | translation reinitiation(GO:0002188) |
| 1.8 | 7.1 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 1.8 | 28.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 1.7 | 6.9 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 1.6 | 8.0 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 1.6 | 6.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 1.5 | 4.5 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 1.3 | 13.3 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 1.1 | 4.6 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 1.0 | 10.3 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 1.0 | 4.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 1.0 | 3.0 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 1.0 | 12.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.9 | 2.8 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.9 | 3.7 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.9 | 3.7 | GO:0035720 | signal transduction downstream of smoothened(GO:0007227) intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.9 | 3.6 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.8 | 3.2 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.8 | 11.7 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.8 | 17.9 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.7 | 7.1 | GO:0072221 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) |
| 0.7 | 2.1 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.6 | 2.3 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.6 | 2.3 | GO:0009189 | dTTP biosynthetic process(GO:0006235) deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.6 | 3.4 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.5 | 4.4 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.5 | 13.8 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.5 | 3.7 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) positive regulation of exit from mitosis(GO:0031536) |
| 0.4 | 1.3 | GO:0060974 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.4 | 1.7 | GO:0060708 | histone H3-K27 acetylation(GO:0043974) leukemia inhibitory factor signaling pathway(GO:0048861) spongiotrophoblast differentiation(GO:0060708) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.4 | 3.6 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.4 | 4.0 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.4 | 3.1 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.4 | 3.9 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.4 | 5.6 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.4 | 7.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.4 | 0.7 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.3 | 12.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.3 | 2.0 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.3 | 2.7 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.3 | 1.7 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.3 | 2.3 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.3 | 2.3 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.3 | 1.5 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.3 | 1.8 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.3 | 1.5 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.3 | 22.1 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.3 | 2.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 6.4 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.2 | 2.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.2 | 2.9 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.2 | 2.8 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 3.8 | GO:0002192 | IRES-dependent translational initiation(GO:0002192) |
| 0.2 | 6.3 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.2 | 2.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 1.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.2 | 1.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.2 | 1.3 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.1 | 1.4 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.1 | 5.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 4.3 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.1 | 1.3 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 0.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 4.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.7 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 2.5 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 0.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.5 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.1 | 0.5 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 2.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 5.8 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 2.7 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.1 | 0.2 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 1.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 5.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.1 | 0.5 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 3.2 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 1.4 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.1 | 0.5 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 2.9 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.1 | 1.0 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 8.5 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 5.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.7 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 2.4 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.8 | GO:0035264 | multicellular organism growth(GO:0035264) |
| 0.0 | 4.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 3.2 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.0 | 2.0 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 1.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 1.7 | GO:0009988 | cell-cell recognition(GO:0009988) |
| 0.0 | 1.0 | GO:0045143 | homologous chromosome segregation(GO:0045143) |
| 0.0 | 0.5 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.9 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.6 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.0 | 1.2 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 1.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.3 | GO:0070985 | TFIIK complex(GO:0070985) |
| 2.5 | 12.4 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 1.6 | 4.7 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.2 | 3.7 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.0 | 3.9 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.9 | 2.8 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.9 | 13.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.9 | 3.6 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.6 | 3.7 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.6 | 3.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.5 | 28.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.5 | 4.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.4 | 4.4 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.4 | 3.0 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.3 | 1.3 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.3 | 4.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.3 | 2.7 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 31.4 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.2 | 2.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 2.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.2 | 2.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 5.1 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.2 | 2.2 | GO:0001673 | male germ cell nucleus(GO:0001673) XY body(GO:0001741) |
| 0.2 | 4.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 13.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 2.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 1.8 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 3.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 2.5 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 2.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.9 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 14.9 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 5.6 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 6.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 1.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 4.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 3.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 3.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.1 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 7.9 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 2.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 8.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 12.1 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 3.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 4.1 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 2.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 5.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 10.4 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 5.9 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.7 | GO:0018995 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 14.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 6.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 6.9 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.7 | GO:0072562 | blood microparticle(GO:0072562) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 8.9 | GO:0070546 | phosphatidylserine decarboxylase activity(GO:0004609) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 2.8 | 28.0 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 2.5 | 12.4 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 2.1 | 12.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 1.6 | 4.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 1.3 | 4.0 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 1.3 | 12.6 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 1.0 | 4.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 1.0 | 6.9 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 1.0 | 3.9 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.9 | 2.8 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.8 | 8.8 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.8 | 3.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.8 | 12.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.7 | 24.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.7 | 2.1 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.7 | 23.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.7 | 2.0 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.6 | 1.3 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.6 | 12.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.6 | 4.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.6 | 3.3 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.5 | 4.4 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.4 | 3.0 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.4 | 10.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.4 | 2.5 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.4 | 2.3 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.3 | 1.3 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.3 | 6.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.3 | 0.9 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.3 | 4.1 | GO:0003993 | acid phosphatase activity(GO:0003993) non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.3 | 1.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 0.7 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.2 | 3.6 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 1.7 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 9.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 2.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 5.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 3.4 | GO:0070016 | gamma-catenin binding(GO:0045295) armadillo repeat domain binding(GO:0070016) |
| 0.2 | 2.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 1.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 2.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 2.0 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 4.9 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.1 | 3.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.8 | GO:0043495 | protein anchor(GO:0043495) dynein intermediate chain binding(GO:0045505) |
| 0.1 | 1.5 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.4 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 4.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 5.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 2.5 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 7.2 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 4.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.3 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 1.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 1.1 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 3.8 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 2.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 4.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 2.3 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 5.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 12.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 2.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.5 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 4.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 3.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.1 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 4.6 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 5.1 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 2.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 1.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 4.1 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 10.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 14.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 12.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 14.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 5.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 7.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 2.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.2 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 6.7 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 6.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 10.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 3.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 2.3 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 1.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 13.8 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.9 | 23.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.9 | 23.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.7 | 17.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.7 | 12.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.5 | 7.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.4 | 10.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.3 | 4.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 4.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 11.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.2 | 5.6 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.2 | 6.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 3.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 2.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 3.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 7.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 3.7 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 3.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 5.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 5.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 4.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 2.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 4.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 4.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 5.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 3.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |