Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
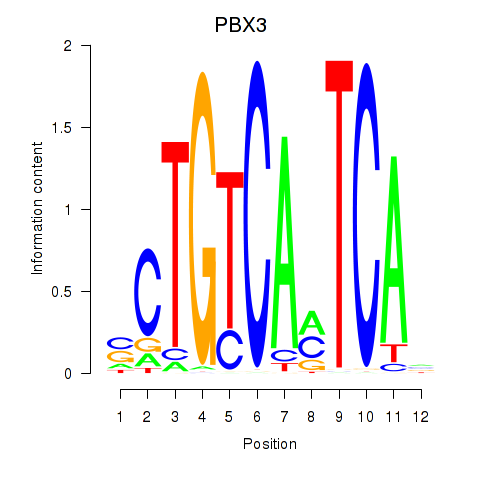
Results for PBX3
Z-value: 1.20
Transcription factors associated with PBX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PBX3
|
ENSG00000167081.12 | PBX homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PBX3 | hg19_v2_chr9_+_128509624_128509658 | -0.17 | 1.4e-02 | Click! |
Activity profile of PBX3 motif
Sorted Z-values of PBX3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PBX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 12.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 3.9 | 11.6 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 2.7 | 21.4 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 2.4 | 14.7 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 2.4 | 14.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 2.4 | 11.8 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 2.2 | 37.2 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 2.1 | 10.5 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 2.0 | 24.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.9 | 20.9 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 1.8 | 14.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 1.5 | 5.9 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 1.4 | 7.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 1.4 | 8.4 | GO:0070417 | cellular response to cold(GO:0070417) |
| 1.4 | 8.4 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 1.3 | 5.2 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 1.3 | 2.5 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 1.2 | 2.5 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) convergent extension involved in axis elongation(GO:0060028) |
| 1.1 | 3.4 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) Spemann organizer formation(GO:0060061) |
| 1.1 | 4.3 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 1.1 | 4.3 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 1.1 | 3.2 | GO:0060364 | frontal suture morphogenesis(GO:0060364) |
| 1.0 | 5.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 1.0 | 3.1 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 1.0 | 5.0 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 1.0 | 3.0 | GO:0090076 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) relaxation of skeletal muscle(GO:0090076) |
| 1.0 | 7.9 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 1.0 | 5.8 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.8 | 10.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.8 | 5.9 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.8 | 3.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.8 | 2.5 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.8 | 1.6 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.8 | 4.0 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.8 | 3.2 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.8 | 2.3 | GO:0021592 | fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 0.7 | 4.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.7 | 11.8 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.7 | 30.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.7 | 4.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.7 | 4.8 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.7 | 8.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.7 | 5.4 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.6 | 15.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.6 | 2.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.6 | 17.4 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.6 | 4.1 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.6 | 4.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.6 | 18.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.6 | 7.9 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.6 | 6.7 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.5 | 21.2 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.5 | 6.5 | GO:0007512 | adult heart development(GO:0007512) |
| 0.5 | 3.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.5 | 28.5 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.5 | 9.6 | GO:0034776 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.5 | 1.9 | GO:2001054 | regulation of mesenchymal cell apoptotic process(GO:2001053) negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.5 | 3.3 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.5 | 1.4 | GO:0072560 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.5 | 3.7 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.5 | 1.4 | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.5 | 2.8 | GO:0071910 | determination of pancreatic left/right asymmetry(GO:0035469) determination of liver left/right asymmetry(GO:0071910) |
| 0.5 | 1.8 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.4 | 2.6 | GO:0021764 | amygdala development(GO:0021764) |
| 0.4 | 4.3 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.4 | 4.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.4 | 2.9 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.4 | 1.7 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.4 | 2.9 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.4 | 7.2 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.4 | 4.6 | GO:0090520 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.4 | 3.7 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.4 | 1.5 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.4 | 0.7 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.4 | 8.0 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.4 | 7.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.4 | 5.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.4 | 1.1 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.3 | 1.4 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.3 | 1.4 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.3 | 1.7 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.3 | 1.3 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.3 | 2.0 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.3 | 1.0 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.3 | 2.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.3 | 13.4 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.3 | 2.8 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.3 | 6.7 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.3 | 19.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.3 | 5.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.3 | 2.6 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.3 | 5.4 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.3 | 1.5 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.2 | 4.7 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.2 | 2.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.2 | 4.4 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.2 | 0.7 | GO:0030497 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.2 | 2.0 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.2 | 1.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 1.0 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.2 | 4.3 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.2 | 1.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 2.7 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.2 | 0.9 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 0.9 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.2 | 3.8 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.2 | 3.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 0.5 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.2 | 1.5 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.2 | 0.7 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.2 | 13.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.2 | 0.8 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.2 | 0.6 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.2 | 4.0 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 0.7 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.1 | 4.9 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 6.1 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.1 | 0.9 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.1 | 9.8 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 1.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 2.6 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 0.4 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 2.4 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 2.0 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 1.4 | GO:0090312 | positive regulation of protein deacetylation(GO:0090312) |
| 0.1 | 3.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 3.2 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 2.3 | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 | 3.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 3.0 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 0.8 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 1.7 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.1 | 0.7 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.1 | 5.4 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 1.0 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 4.1 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.1 | 0.5 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 3.1 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 8.8 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 1.4 | GO:1902083 | cellular response to hydroperoxide(GO:0071447) negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 2.4 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 0.8 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 2.7 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 5.1 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.2 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 2.8 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.1 | 0.6 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 1.2 | GO:0060322 | head development(GO:0060322) |
| 0.1 | 2.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 2.1 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 0.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.6 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 5.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 1.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 6.0 | GO:0060173 | appendage development(GO:0048736) limb development(GO:0060173) |
| 0.1 | 2.2 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.1 | 7.4 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.1 | 2.5 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 3.1 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 1.8 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.1 | 6.4 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.1 | 7.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.8 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.1 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.0 | 2.8 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 2.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.4 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.2 | GO:0044108 | cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) |
| 0.0 | 4.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.6 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 3.2 | GO:0046323 | glucose import(GO:0046323) |
| 0.0 | 0.3 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.2 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.0 | 0.7 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 | 2.5 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.0 | 24.9 | GO:0007417 | central nervous system development(GO:0007417) |
| 0.0 | 0.4 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 1.4 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 1.7 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 2.1 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 1.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 1.1 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 | 3.5 | GO:0007601 | visual perception(GO:0007601) sensory perception of light stimulus(GO:0050953) |
| 0.0 | 2.4 | GO:0061572 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.0 | 1.6 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 2.9 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 2.7 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 1.5 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 0.7 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.3 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.3 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.6 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 20.1 | GO:0070695 | FHF complex(GO:0070695) |
| 3.5 | 14.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 3.1 | 21.4 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 2.5 | 15.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 2.0 | 5.9 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 1.7 | 24.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.7 | 6.9 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 1.4 | 4.3 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 1.4 | 4.3 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.2 | 17.4 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 1.1 | 11.5 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 1.1 | 14.7 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 1.1 | 7.9 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 1.1 | 3.3 | GO:0060187 | cell pole(GO:0060187) |
| 1.0 | 11.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.7 | 12.0 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.6 | 3.0 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.6 | 2.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.6 | 5.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.6 | 10.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.5 | 1.6 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.5 | 4.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.4 | 0.8 | GO:0098798 | mitochondrial protein complex(GO:0098798) |
| 0.4 | 4.6 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.4 | 1.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.4 | 2.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.3 | 1.3 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.3 | 4.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 6.8 | GO:0010369 | chromocenter(GO:0010369) |
| 0.3 | 0.9 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.3 | 3.4 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.3 | 77.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.3 | 9.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.3 | 2.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 2.0 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.2 | 10.1 | GO:0030496 | midbody(GO:0030496) |
| 0.2 | 8.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 1.0 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.2 | 2.6 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 1.2 | GO:0044308 | axonal spine(GO:0044308) |
| 0.2 | 4.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 5.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 2.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 4.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 3.0 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.2 | 20.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.2 | 13.6 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 2.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.2 | 2.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.2 | 6.0 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 5.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 5.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 6.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 9.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 2.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 2.0 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 3.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.6 | GO:0098827 | terminal cisterna(GO:0014802) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 2.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 6.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 0.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 3.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 7.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 4.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 1.5 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 3.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 5.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.7 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 1.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.6 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 2.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 4.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.0 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 9.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 1.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 4.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.7 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.7 | GO:0005657 | replication fork(GO:0005657) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.6 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 2.9 | 34.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 2.0 | 28.7 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 2.0 | 6.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 2.0 | 7.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 1.8 | 5.4 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 1.5 | 21.4 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 1.4 | 4.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 1.3 | 5.0 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 1.2 | 12.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 1.0 | 1.0 | GO:0016774 | phosphoglycerate kinase activity(GO:0004618) phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 1.0 | 2.9 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.9 | 4.5 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.9 | 2.7 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.9 | 7.9 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.8 | 5.9 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.8 | 3.3 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.8 | 2.5 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.8 | 4.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.8 | 4.8 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 0.8 | 3.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.8 | 7.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.7 | 13.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.7 | 4.3 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.7 | 6.4 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.7 | 9.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.7 | 2.7 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.6 | 2.6 | GO:0052812 | phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.6 | 10.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.6 | 17.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.6 | 6.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.6 | 15.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.6 | 4.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.6 | 1.7 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.5 | 4.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.5 | 6.4 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.5 | 4.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.5 | 3.3 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.5 | 3.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.5 | 3.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.4 | 8.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.4 | 29.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.4 | 4.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.4 | 1.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.4 | 2.9 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.3 | 1.4 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.3 | 1.7 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.3 | 2.7 | GO:0005497 | androgen binding(GO:0005497) |
| 0.3 | 4.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.3 | 4.3 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.3 | 2.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 4.0 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.3 | 22.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 6.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 2.8 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.3 | 1.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 5.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 1.0 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.2 | 2.8 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 5.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.2 | 2.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 4.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 6.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 3.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 1.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.2 | 10.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 2.8 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.2 | 1.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.2 | 13.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.2 | 45.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.2 | 26.6 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.2 | 2.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.2 | 0.9 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.2 | 2.0 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 3.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.2 | 4.0 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.2 | 15.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 1.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 0.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 0.5 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.2 | 0.7 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.2 | 4.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 5.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 1.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 1.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 4.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 4.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 2.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 9.5 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 1.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 2.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 14.2 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 1.0 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 2.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.8 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 1.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 3.0 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 13.7 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.1 | 1.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 3.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 3.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 4.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 2.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 4.3 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.1 | 1.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.8 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.1 | 28.2 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.6 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 2.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 1.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 3.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 261.9 | GO:0046872 | metal ion binding(GO:0046872) |
| 0.1 | 1.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.3 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 1.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 2.0 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.0 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.6 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 2.9 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 4.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 1.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 2.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 1.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 2.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 15.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.5 | 11.6 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.4 | 24.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 10.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.3 | 6.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.3 | 9.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 7.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 3.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 3.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.2 | 12.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 16.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 0.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 3.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 4.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 5.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.7 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 2.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 10.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 1.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.0 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 3.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 2.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 1.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 3.8 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 2.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.6 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 3.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 4.2 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.9 | 24.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.5 | 12.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.5 | 26.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.4 | 10.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.4 | 11.6 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.4 | 6.9 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.3 | 10.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.3 | 10.8 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.3 | 8.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.3 | 29.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.3 | 9.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.3 | 5.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.3 | 10.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.3 | 113.0 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.2 | 7.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 4.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 5.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.2 | 3.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 8.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.2 | 2.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 3.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.2 | 4.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 2.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 7.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 9.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.2 | 1.6 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 2.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 4.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 4.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 9.5 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.1 | 4.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 2.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 5.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 3.0 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 1.5 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 3.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 4.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.6 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 1.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 2.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 0.8 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 2.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 3.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 2.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.4 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |