Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
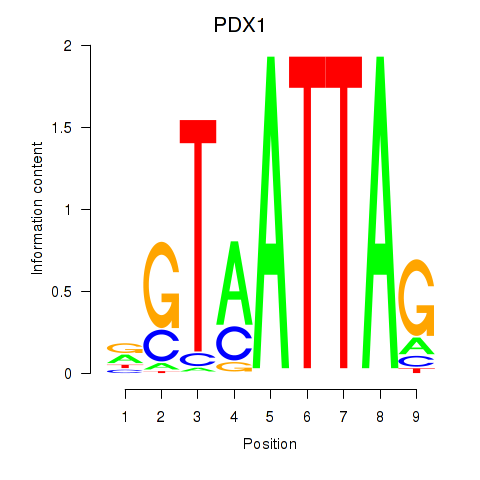
Results for PDX1
Z-value: 0.78
Transcription factors associated with PDX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PDX1
|
ENSG00000139515.5 | pancreatic and duodenal homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PDX1 | hg19_v2_chr13_+_28494130_28494168 | -0.20 | 3.1e-03 | Click! |
Activity profile of PDX1 motif
Sorted Z-values of PDX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_75080883 | 31.98 |
ENST00000567571.1
|
CSK
|
c-src tyrosine kinase |
| chr13_-_46716969 | 23.85 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr15_-_55563072 | 16.12 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr15_-_55562479 | 15.87 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr1_+_198608146 | 15.36 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr6_-_32157947 | 14.35 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr13_-_41593425 | 12.85 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chrX_-_20236970 | 12.73 |
ENST00000379548.4
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr5_-_134735568 | 12.30 |
ENST00000510038.1
ENST00000304332.4 |
H2AFY
|
H2A histone family, member Y |
| chr15_-_55562582 | 12.27 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr4_-_103749179 | 12.16 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_-_103749205 | 12.16 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr7_+_116660246 | 10.78 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr18_+_57567180 | 9.82 |
ENST00000316660.6
ENST00000269518.9 |
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr12_+_50144381 | 9.07 |
ENST00000552370.1
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr12_+_28410128 | 9.03 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr2_+_90060377 | 8.62 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr1_+_174844645 | 8.57 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr2_-_136678123 | 8.55 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr6_+_34204642 | 8.49 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr2_-_89459813 | 8.08 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chr1_-_68698197 | 8.07 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr1_+_158979792 | 7.85 |
ENST00000359709.3
ENST00000430894.2 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr7_-_25268104 | 7.81 |
ENST00000222674.2
|
NPVF
|
neuropeptide VF precursor |
| chr5_+_135394840 | 7.18 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr18_+_3447572 | 6.73 |
ENST00000548489.2
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr6_-_25042231 | 6.56 |
ENST00000510784.2
|
FAM65B
|
family with sequence similarity 65, member B |
| chr8_-_42234745 | 6.21 |
ENST00000220812.2
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr5_-_142780280 | 5.78 |
ENST00000424646.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr10_+_11047259 | 5.77 |
ENST00000379261.4
ENST00000416382.2 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr15_+_58430368 | 5.61 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr17_-_27418537 | 5.48 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr12_+_56325812 | 5.47 |
ENST00000394147.1
ENST00000551156.1 ENST00000553783.1 ENST00000557080.1 ENST00000432422.3 ENST00000556001.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr1_+_45241109 | 5.12 |
ENST00000396651.3
|
RPS8
|
ribosomal protein S8 |
| chr15_+_58430567 | 5.11 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr3_-_39321512 | 5.09 |
ENST00000399220.2
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1 |
| chr21_-_15918618 | 4.82 |
ENST00000400564.1
ENST00000400566.1 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr2_-_61697862 | 4.73 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr3_+_136649311 | 4.50 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr13_-_31191642 | 4.46 |
ENST00000405805.1
|
HMGB1
|
high mobility group box 1 |
| chr1_-_151965048 | 4.39 |
ENST00000368809.1
|
S100A10
|
S100 calcium binding protein A10 |
| chr5_-_141249154 | 4.35 |
ENST00000357517.5
ENST00000536585.1 |
PCDH1
|
protocadherin 1 |
| chr1_-_1709845 | 4.29 |
ENST00000341426.5
ENST00000344463.4 |
NADK
|
NAD kinase |
| chr4_-_103749313 | 4.20 |
ENST00000394803.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr12_+_49621658 | 4.19 |
ENST00000541364.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr3_-_57233966 | 4.12 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr16_-_30457048 | 3.96 |
ENST00000500504.2
ENST00000542752.1 |
SEPHS2
|
selenophosphate synthetase 2 |
| chr3_-_141747950 | 3.94 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr17_-_77924627 | 3.86 |
ENST00000572862.1
ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16
|
TBC1 domain family, member 16 |
| chr1_-_1710229 | 3.79 |
ENST00000341991.3
|
NADK
|
NAD kinase |
| chr8_+_9953214 | 3.78 |
ENST00000382490.5
|
MSRA
|
methionine sulfoxide reductase A |
| chr2_-_145277569 | 3.62 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr15_+_80351910 | 3.62 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr14_+_72052983 | 3.61 |
ENST00000358550.2
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr1_+_68150744 | 3.57 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr4_-_103749105 | 3.47 |
ENST00000394801.4
ENST00000394804.2 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr8_+_9953061 | 3.46 |
ENST00000522907.1
ENST00000528246.1 |
MSRA
|
methionine sulfoxide reductase A |
| chr4_-_103748880 | 3.42 |
ENST00000453744.2
ENST00000349311.8 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr18_-_33709268 | 3.38 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr7_+_100860949 | 3.25 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr12_-_86650077 | 3.19 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr13_+_102104980 | 3.16 |
ENST00000545560.2
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr12_-_54813229 | 3.13 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr11_-_8290263 | 3.02 |
ENST00000428101.2
|
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr10_+_114710211 | 3.00 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr6_-_109702885 | 3.00 |
ENST00000504373.1
|
CD164
|
CD164 molecule, sialomucin |
| chrX_-_110655306 | 2.96 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr1_+_36621529 | 2.96 |
ENST00000316156.4
|
MAP7D1
|
MAP7 domain containing 1 |
| chr7_-_115670804 | 2.90 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr9_+_125133315 | 2.85 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr20_+_30697298 | 2.80 |
ENST00000398022.2
|
TM9SF4
|
transmembrane 9 superfamily protein member 4 |
| chr10_-_48416849 | 2.75 |
ENST00000249598.1
|
GDF2
|
growth differentiation factor 2 |
| chr19_+_13049413 | 2.73 |
ENST00000316448.5
ENST00000588454.1 |
CALR
|
calreticulin |
| chr2_+_109204743 | 2.73 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr19_+_11071546 | 2.72 |
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr7_-_115670792 | 2.72 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr1_-_212965104 | 2.68 |
ENST00000422588.2
ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1
|
NSL1, MIS12 kinetochore complex component |
| chr18_+_32173276 | 2.61 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr18_+_59000815 | 2.51 |
ENST00000262717.4
|
CDH20
|
cadherin 20, type 2 |
| chr4_+_169013666 | 2.48 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr9_+_125132803 | 2.44 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr14_-_78083112 | 2.44 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr3_+_108541608 | 2.33 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr1_+_101003687 | 2.30 |
ENST00000315033.4
|
GPR88
|
G protein-coupled receptor 88 |
| chr11_-_102496063 | 2.29 |
ENST00000260228.2
|
MMP20
|
matrix metallopeptidase 20 |
| chr12_-_32908809 | 2.18 |
ENST00000324868.8
|
YARS2
|
tyrosyl-tRNA synthetase 2, mitochondrial |
| chr4_+_151503077 | 2.15 |
ENST00000317605.4
|
MAB21L2
|
mab-21-like 2 (C. elegans) |
| chr4_-_66536057 | 2.07 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr5_-_140998616 | 2.05 |
ENST00000389054.3
ENST00000398562.2 ENST00000389057.5 ENST00000398566.3 ENST00000398557.4 ENST00000253811.6 |
DIAPH1
|
diaphanous-related formin 1 |
| chr19_+_50016610 | 2.04 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chrX_-_106243451 | 2.01 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr3_+_108541545 | 2.01 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr18_+_55888767 | 2.00 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr7_+_50348268 | 2.00 |
ENST00000438033.1
ENST00000439701.1 |
IKZF1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr7_+_129015484 | 1.85 |
ENST00000490911.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr15_+_59279851 | 1.84 |
ENST00000348370.4
ENST00000434298.1 ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chr17_+_60536002 | 1.83 |
ENST00000582809.1
|
TLK2
|
tousled-like kinase 2 |
| chr4_+_30721968 | 1.82 |
ENST00000361762.2
|
PCDH7
|
protocadherin 7 |
| chr4_-_116034979 | 1.78 |
ENST00000264363.2
|
NDST4
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
| chr6_-_38670897 | 1.78 |
ENST00000373365.4
|
GLO1
|
glyoxalase I |
| chr4_-_39979576 | 1.74 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr3_-_55521323 | 1.73 |
ENST00000264634.4
|
WNT5A
|
wingless-type MMTV integration site family, member 5A |
| chr14_+_22985251 | 1.69 |
ENST00000390510.1
|
TRAJ27
|
T cell receptor alpha joining 27 |
| chr3_+_69985734 | 1.63 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr13_-_99667960 | 1.61 |
ENST00000448493.2
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr7_-_81399355 | 1.59 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr15_+_80351977 | 1.51 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr3_-_108248169 | 1.44 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr17_-_6983594 | 1.43 |
ENST00000571664.1
ENST00000254868.4 |
CLEC10A
|
C-type lectin domain family 10, member A |
| chr15_-_31393910 | 1.41 |
ENST00000397795.2
ENST00000256552.6 ENST00000559179.1 |
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr1_+_159272111 | 1.30 |
ENST00000368114.1
|
FCER1A
|
Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide |
| chr4_+_144354644 | 1.29 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr1_-_2458026 | 1.27 |
ENST00000435556.3
ENST00000378466.3 |
PANK4
|
pantothenate kinase 4 |
| chr4_-_66536196 | 1.27 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr5_+_156712372 | 1.27 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr7_-_81399438 | 1.25 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr14_-_69261310 | 1.25 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr4_-_41750922 | 1.23 |
ENST00000226382.2
|
PHOX2B
|
paired-like homeobox 2b |
| chr1_-_151431647 | 1.22 |
ENST00000368863.2
ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ
|
pogo transposable element with ZNF domain |
| chr20_+_62795827 | 1.22 |
ENST00000328439.1
ENST00000536311.1 |
MYT1
|
myelin transcription factor 1 |
| chr18_+_28898052 | 1.20 |
ENST00000257192.4
|
DSG1
|
desmoglein 1 |
| chr9_-_136933134 | 1.16 |
ENST00000303407.7
|
BRD3
|
bromodomain containing 3 |
| chr11_-_8285405 | 1.16 |
ENST00000335790.3
ENST00000534484.1 |
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr14_-_36988882 | 1.15 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr12_-_52967600 | 1.11 |
ENST00000549343.1
ENST00000305620.2 |
KRT74
|
keratin 74 |
| chr4_-_123377880 | 1.04 |
ENST00000226730.4
|
IL2
|
interleukin 2 |
| chr2_+_45168875 | 0.99 |
ENST00000260653.3
|
SIX3
|
SIX homeobox 3 |
| chr9_+_136501478 | 0.95 |
ENST00000393056.2
ENST00000263611.2 |
DBH
|
dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| chr12_-_86650045 | 0.95 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr12_-_14849470 | 0.93 |
ENST00000261170.3
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr5_-_9630463 | 0.92 |
ENST00000382492.2
|
TAS2R1
|
taste receptor, type 2, member 1 |
| chr17_-_6983550 | 0.89 |
ENST00000576617.1
ENST00000416562.2 |
CLEC10A
|
C-type lectin domain family 10, member A |
| chrX_+_144908928 | 0.86 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr18_+_29027696 | 0.86 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chrX_-_18690210 | 0.74 |
ENST00000379984.3
|
RS1
|
retinoschisin 1 |
| chr12_-_53171128 | 0.73 |
ENST00000332411.2
|
KRT76
|
keratin 76 |
| chr6_+_167704798 | 0.69 |
ENST00000230256.3
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr6_-_31107127 | 0.67 |
ENST00000259845.4
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chr2_-_190927447 | 0.66 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr20_+_52105495 | 0.62 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chrX_-_64196376 | 0.62 |
ENST00000447788.2
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr11_-_102709441 | 0.62 |
ENST00000434103.1
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr15_+_48498480 | 0.60 |
ENST00000380993.3
ENST00000396577.3 |
SLC12A1
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr4_-_119759795 | 0.56 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chrX_-_64196307 | 0.55 |
ENST00000545618.1
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr7_-_81399329 | 0.54 |
ENST00000453411.1
ENST00000444829.2 |
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr3_+_69985792 | 0.54 |
ENST00000531774.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr5_-_134871639 | 0.54 |
ENST00000314744.4
|
NEUROG1
|
neurogenin 1 |
| chr21_-_35899113 | 0.52 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr8_+_22424551 | 0.47 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr7_-_151217166 | 0.46 |
ENST00000496004.1
|
RHEB
|
Ras homolog enriched in brain |
| chr1_-_94586651 | 0.44 |
ENST00000535735.1
ENST00000370225.3 |
ABCA4
|
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr6_+_150920999 | 0.43 |
ENST00000367328.1
ENST00000367326.1 |
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr11_-_36619771 | 0.42 |
ENST00000311485.3
ENST00000527033.1 ENST00000532616.1 |
RAG2
|
recombination activating gene 2 |
| chr4_-_139163491 | 0.38 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chrX_-_64196351 | 0.36 |
ENST00000374839.3
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr4_-_70518941 | 0.33 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr1_+_199996702 | 0.30 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr19_+_641178 | 0.29 |
ENST00000166133.3
|
FGF22
|
fibroblast growth factor 22 |
| chr6_+_167704838 | 0.25 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr12_-_86230315 | 0.20 |
ENST00000361228.3
|
RASSF9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr1_-_152386732 | 0.20 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chr6_+_13272904 | 0.20 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr7_-_81399411 | 0.18 |
ENST00000423064.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr5_+_169532896 | 0.15 |
ENST00000306268.6
ENST00000449804.2 |
FOXI1
|
forkhead box I1 |
| chr6_+_29079668 | 0.13 |
ENST00000377169.1
|
OR2J3
|
olfactory receptor, family 2, subfamily J, member 3 |
| chr4_-_155533787 | 0.13 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr15_-_72563585 | 0.10 |
ENST00000287196.9
ENST00000260376.7 |
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr6_-_33160231 | 0.07 |
ENST00000395194.1
ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2
|
collagen, type XI, alpha 2 |
| chr20_+_43029911 | 0.03 |
ENST00000443598.2
ENST00000316099.4 ENST00000415691.2 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr12_-_51611477 | 0.01 |
ENST00000389243.4
|
POU6F1
|
POU class 6 homeobox 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PDX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 44.3 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 6.4 | 32.0 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 4.1 | 12.3 | GO:1901837 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 3.1 | 15.4 | GO:0048539 | immunoglobulin biosynthetic process(GO:0002378) bone marrow development(GO:0048539) |
| 2.8 | 8.5 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 2.4 | 7.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 2.1 | 10.7 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) urea transmembrane transport(GO:0071918) purine nucleobase transmembrane transport(GO:1904823) |
| 2.1 | 8.5 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 1.7 | 5.1 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 1.5 | 9.1 | GO:0060701 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 1.5 | 4.5 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 1.5 | 23.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 1.3 | 8.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 1.3 | 4.0 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 1.3 | 5.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 1.1 | 4.5 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 1.0 | 6.2 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 1.0 | 9.8 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.9 | 3.6 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.8 | 7.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.8 | 7.8 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.8 | 35.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.8 | 9.0 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.7 | 5.8 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.7 | 2.9 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.6 | 2.4 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.6 | 1.7 | GO:1904933 | cardiac right atrium morphogenesis(GO:0003213) negative regulation of melanin biosynthetic process(GO:0048022) positive regulation of anagen(GO:0051885) mediolateral intercalation(GO:0060031) cervix development(GO:0060067) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) negative regulation of secondary metabolite biosynthetic process(GO:1900377) regulation of cell proliferation in midbrain(GO:1904933) |
| 0.6 | 2.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.5 | 2.7 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.5 | 4.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.5 | 15.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.5 | 12.7 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.5 | 12.9 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.5 | 2.7 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.4 | 3.6 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.4 | 1.3 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.4 | 1.2 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.4 | 1.2 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.4 | 2.0 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.4 | 4.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.3 | 3.4 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.3 | 2.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.3 | 5.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.3 | 2.0 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.3 | 1.8 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.3 | 3.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.3 | 1.8 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.2 | 2.8 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 4.4 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.2 | 1.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.2 | 1.8 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 | 5.5 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.2 | 0.4 | GO:0002329 | pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.2 | 2.3 | GO:0070170 | regulation of tooth mineralization(GO:0070170) |
| 0.2 | 1.8 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 | 5.1 | GO:0072663 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.2 | 4.7 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.2 | 3.6 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.2 | 3.3 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.2 | 0.5 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.2 | 2.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.2 | 0.7 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) negative regulation of skeletal muscle cell proliferation(GO:0014859) regulation of skeletal muscle tissue growth(GO:0048631) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.2 | 7.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.2 | 3.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.2 | 1.0 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.2 | 1.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.2 | 5.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 0.3 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.2 | 4.8 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 16.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 1.8 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 2.7 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 5.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 16.7 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 2.2 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 0.7 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 3.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.9 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 1.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.7 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.1 | 2.7 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of adherens junction organization(GO:1903393) |
| 0.1 | 2.6 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 1.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 2.2 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 0.6 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 1.5 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 1.3 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 1.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 6.9 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.2 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.1 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 1.0 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 4.2 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 2.7 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.4 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 2.0 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.5 | GO:0043410 | positive regulation of MAPK cascade(GO:0043410) |
| 0.0 | 0.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 4.4 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 3.9 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.1 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.0 | 0.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 32.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 2.1 | 12.3 | GO:0001740 | Barr body(GO:0001740) |
| 1.2 | 23.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.8 | 3.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.8 | 8.5 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.6 | 2.4 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.5 | 2.7 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.5 | 2.9 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.4 | 8.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.4 | 1.4 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.3 | 2.7 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.3 | 4.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 7.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 34.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.2 | 5.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.2 | 3.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 4.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 2.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 5.1 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.1 | 2.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 19.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 3.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.7 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 1.0 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 2.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 5.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 2.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 5.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 18.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 1.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 7.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 29.3 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 1.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 9.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 3.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 3.0 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 1.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.2 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 8.6 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 9.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.7 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 1.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 4.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 8.1 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 7.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 3.4 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 52.9 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 13.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 4.8 | GO:0031966 | mitochondrial membrane(GO:0031966) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.7 | GO:0015265 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 1.7 | 8.5 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 1.7 | 44.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 1.7 | 5.1 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 1.5 | 9.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 1.4 | 7.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 1.4 | 12.3 | GO:0000182 | rDNA binding(GO:0000182) |
| 1.3 | 5.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 1.3 | 4.0 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 1.3 | 32.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 1.2 | 5.8 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.9 | 4.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.8 | 2.4 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.8 | 35.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.7 | 4.5 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.6 | 1.8 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.6 | 8.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.6 | 4.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.5 | 7.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.5 | 14.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.5 | 2.0 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.5 | 13.6 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.3 | 2.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 3.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 1.3 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.3 | 1.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.3 | 1.8 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.3 | 1.8 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.3 | 4.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.3 | 3.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.3 | 12.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.3 | 2.0 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.2 | 6.7 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 4.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 1.8 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 2.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 1.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 3.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 1.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 1.0 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 5.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 5.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 5.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 3.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 4.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 4.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 1.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.9 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 1.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 0.4 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 12.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 16.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 10.4 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 3.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 2.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 3.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 2.2 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 29.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 13.8 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.6 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 1.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 4.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.6 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 4.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 3.7 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 2.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 5.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 4.1 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 7.0 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 2.1 | GO:0008017 | microtubule binding(GO:0008017) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 35.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.6 | 51.9 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.4 | 12.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.3 | 13.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 15.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.2 | 3.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.2 | 11.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 18.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 5.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 5.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 2.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 7.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 4.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 3.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 3.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 3.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 3.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 6.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 3.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 4.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 3.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 4.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 47.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 1.7 | 44.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 1.0 | 35.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.9 | 8.5 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.8 | 10.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.4 | 9.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 12.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 8.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 3.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 6.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 2.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 4.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 3.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 4.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 3.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 4.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 4.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 2.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 2.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 5.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 2.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 5.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 7.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 5.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 3.1 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 5.3 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 1.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 1.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.0 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 2.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 1.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |