Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
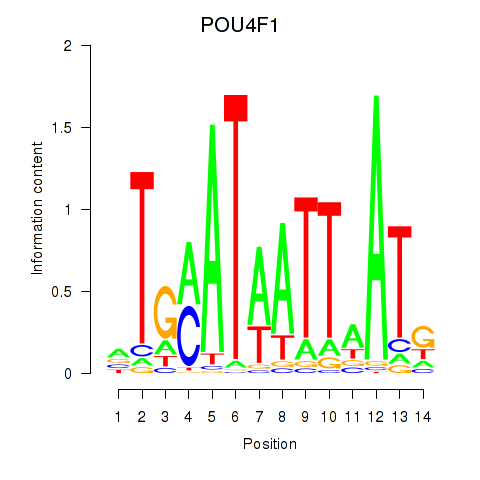
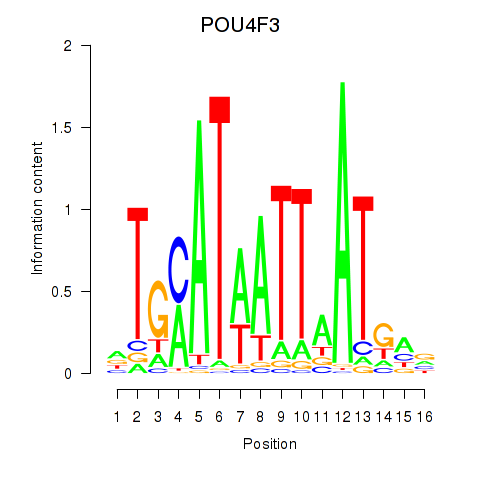
Results for POU4F1_POU4F3
Z-value: 0.71
Transcription factors associated with POU4F1_POU4F3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU4F1
|
ENSG00000152192.6 | POU class 4 homeobox 1 |
|
POU4F3
|
ENSG00000091010.4 | POU class 4 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU4F1 | hg19_v2_chr13_-_79177673_79177701 | 0.30 | 5.7e-06 | Click! |
| POU4F3 | hg19_v2_chr5_+_145718587_145718607 | 0.10 | 1.5e-01 | Click! |
Activity profile of POU4F1_POU4F3 motif
Sorted Z-values of POU4F1_POU4F3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_130554803 | 25.48 |
ENST00000535487.1
|
RP11-474D1.2
|
RP11-474D1.2 |
| chr12_-_91574142 | 23.94 |
ENST00000547937.1
|
DCN
|
decorin |
| chr2_-_88285309 | 15.12 |
ENST00000420840.2
|
RGPD2
|
RANBP2-like and GRIP domain containing 2 |
| chr19_-_18632861 | 14.89 |
ENST00000262809.4
|
ELL
|
elongation factor RNA polymerase II |
| chr7_-_100026280 | 13.15 |
ENST00000360951.4
ENST00000398027.2 ENST00000324725.6 ENST00000472716.1 |
ZCWPW1
|
zinc finger, CW type with PWWP domain 1 |
| chr2_+_87135076 | 13.07 |
ENST00000409776.2
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr22_+_41777927 | 10.33 |
ENST00000266304.4
|
TEF
|
thyrotrophic embryonic factor |
| chr15_+_74422585 | 10.07 |
ENST00000561740.1
ENST00000435464.1 ENST00000453268.2 |
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr12_-_6798616 | 9.16 |
ENST00000355772.4
ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384
|
zinc finger protein 384 |
| chr12_-_42631529 | 8.85 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr12_-_6798410 | 8.05 |
ENST00000361959.3
ENST00000436774.2 ENST00000544482.1 |
ZNF384
|
zinc finger protein 384 |
| chr12_-_6798523 | 8.04 |
ENST00000319770.3
|
ZNF384
|
zinc finger protein 384 |
| chr18_-_52989217 | 8.04 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr14_+_22977587 | 7.50 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chr14_+_101297740 | 6.87 |
ENST00000555928.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr5_+_129083772 | 6.27 |
ENST00000564719.1
|
KIAA1024L
|
KIAA1024-like |
| chr12_-_91546926 | 5.83 |
ENST00000550758.1
|
DCN
|
decorin |
| chr14_-_21270995 | 5.73 |
ENST00000555698.1
ENST00000397970.4 ENST00000340900.3 |
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr16_+_15596123 | 5.00 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr9_+_34458771 | 4.93 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr14_-_21270561 | 4.71 |
ENST00000412779.2
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chrX_-_138724994 | 4.30 |
ENST00000536274.1
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr6_-_52859046 | 4.09 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr4_-_16085314 | 3.95 |
ENST00000510224.1
|
PROM1
|
prominin 1 |
| chr17_-_40540484 | 3.77 |
ENST00000588969.1
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr7_-_100425112 | 3.75 |
ENST00000358173.3
|
EPHB4
|
EPH receptor B4 |
| chr4_+_102711764 | 3.72 |
ENST00000322953.4
ENST00000428908.1 |
BANK1
|
B-cell scaffold protein with ankyrin repeats 1 |
| chr2_+_87144738 | 3.68 |
ENST00000559485.1
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr4_+_106631966 | 3.60 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr5_-_134871639 | 3.39 |
ENST00000314744.4
|
NEUROG1
|
neurogenin 1 |
| chr4_+_89513574 | 3.38 |
ENST00000402738.1
ENST00000431413.1 ENST00000422770.1 ENST00000407637.1 |
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr1_+_160336851 | 3.31 |
ENST00000302101.5
|
NHLH1
|
nescient helix loop helix 1 |
| chr10_+_104613980 | 3.28 |
ENST00000339834.5
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr4_-_25032501 | 3.26 |
ENST00000382114.4
|
LGI2
|
leucine-rich repeat LGI family, member 2 |
| chr2_-_183387283 | 3.25 |
ENST00000435564.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr4_+_74269956 | 3.08 |
ENST00000295897.4
ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB
|
albumin |
| chr3_-_194072019 | 3.07 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chr6_+_126102292 | 3.06 |
ENST00000368357.3
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr14_-_101036119 | 2.95 |
ENST00000355173.2
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr2_-_183387064 | 2.84 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr4_-_16085340 | 2.82 |
ENST00000508167.1
|
PROM1
|
prominin 1 |
| chr6_-_136847099 | 2.82 |
ENST00000438100.2
|
MAP7
|
microtubule-associated protein 7 |
| chr22_+_40322623 | 2.78 |
ENST00000399090.2
|
GRAP2
|
GRB2-related adaptor protein 2 |
| chr3_+_35722487 | 2.72 |
ENST00000441454.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr2_-_183387430 | 2.70 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr1_-_95538492 | 2.69 |
ENST00000370205.5
|
ALG14
|
ALG14, UDP-N-acetylglucosaminyltransferase subunit |
| chr17_-_29641084 | 2.66 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr22_+_40322595 | 2.66 |
ENST00000420971.1
ENST00000544756.1 |
GRAP2
|
GRB2-related adaptor protein 2 |
| chr22_-_43010928 | 2.62 |
ENST00000348657.2
ENST00000252115.5 |
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chrX_+_41583408 | 2.61 |
ENST00000302548.4
|
GPR82
|
G protein-coupled receptor 82 |
| chr5_-_41261540 | 2.60 |
ENST00000263413.3
|
C6
|
complement component 6 |
| chr1_-_33430286 | 2.57 |
ENST00000373456.7
ENST00000356990.5 ENST00000235150.4 |
RNF19B
|
ring finger protein 19B |
| chr7_-_140482926 | 2.56 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr14_-_65409438 | 2.56 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr19_+_54466179 | 2.54 |
ENST00000270458.2
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr18_+_29027696 | 2.52 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr3_-_149470229 | 2.42 |
ENST00000473414.1
|
COMMD2
|
COMM domain containing 2 |
| chr6_-_44281043 | 2.41 |
ENST00000244571.4
|
AARS2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr2_-_26781521 | 2.39 |
ENST00000403946.3
ENST00000272371.2 |
OTOF
|
otoferlin |
| chr3_-_18480260 | 2.36 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr1_+_50574585 | 2.32 |
ENST00000371824.1
ENST00000371823.4 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr1_-_160549235 | 2.32 |
ENST00000368054.3
ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84
|
CD84 molecule |
| chrX_-_138724677 | 2.32 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr9_-_123812542 | 2.31 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr1_+_198608146 | 2.28 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr1_+_145292806 | 2.24 |
ENST00000448873.2
|
NBPF10
|
neuroblastoma breakpoint family, member 10 |
| chr19_-_53696587 | 2.22 |
ENST00000396424.3
ENST00000600412.1 |
ZNF665
|
zinc finger protein 665 |
| chr13_+_52586517 | 2.18 |
ENST00000523764.1
ENST00000521508.1 |
ALG11
|
ALG11, alpha-1,2-mannosyltransferase |
| chrX_+_135279179 | 2.16 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr16_+_33204980 | 2.10 |
ENST00000561509.1
|
TP53TG3C
|
TP53 target 3C |
| chr15_-_65426174 | 2.10 |
ENST00000204549.4
|
PDCD7
|
programmed cell death 7 |
| chr1_+_66458072 | 2.01 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr16_+_6533380 | 1.98 |
ENST00000552089.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr1_+_206808868 | 1.84 |
ENST00000367109.2
|
DYRK3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr1_+_206808918 | 1.82 |
ENST00000367108.3
|
DYRK3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr17_+_29664830 | 1.82 |
ENST00000444181.2
ENST00000417592.2 |
NF1
|
neurofibromin 1 |
| chr12_-_102224457 | 1.81 |
ENST00000549165.1
ENST00000549940.1 ENST00000392919.4 |
GNPTAB
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr12_+_49740700 | 1.80 |
ENST00000549441.2
ENST00000395069.3 |
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr19_-_36304201 | 1.79 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr1_-_216978709 | 1.72 |
ENST00000360012.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr21_-_37914898 | 1.71 |
ENST00000399136.1
|
CLDN14
|
claudin 14 |
| chrX_+_135278908 | 1.71 |
ENST00000539015.1
ENST00000370683.1 |
FHL1
|
four and a half LIM domains 1 |
| chr15_-_20193370 | 1.63 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr3_-_52860850 | 1.61 |
ENST00000441637.2
|
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chrX_-_32173579 | 1.60 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr16_+_48278178 | 1.55 |
ENST00000285737.4
ENST00000535754.1 |
LONP2
|
lon peptidase 2, peroxisomal |
| chr7_+_100026406 | 1.54 |
ENST00000414441.1
|
MEPCE
|
methylphosphate capping enzyme |
| chr9_-_95298314 | 1.54 |
ENST00000344604.5
ENST00000375540.1 |
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr12_+_110011571 | 1.48 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr17_+_18012020 | 1.40 |
ENST00000205890.5
|
MYO15A
|
myosin XVA |
| chr7_+_134528635 | 1.40 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr2_+_204732487 | 1.39 |
ENST00000302823.3
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr9_-_215744 | 1.36 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chrX_-_65253506 | 1.34 |
ENST00000427538.1
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr2_-_183106641 | 1.34 |
ENST00000346717.4
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr1_-_156470515 | 1.28 |
ENST00000340875.5
ENST00000368240.2 ENST00000353795.3 |
MEF2D
|
myocyte enhancer factor 2D |
| chr3_-_58613323 | 1.28 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr2_-_101034070 | 1.24 |
ENST00000264249.3
|
CHST10
|
carbohydrate sulfotransferase 10 |
| chr1_+_67773044 | 1.23 |
ENST00000262345.1
ENST00000371000.1 |
IL12RB2
|
interleukin 12 receptor, beta 2 |
| chr10_+_135207598 | 1.22 |
ENST00000477902.2
|
MTG1
|
mitochondrial ribosome-associated GTPase 1 |
| chr5_-_135290705 | 1.22 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr7_-_115670792 | 1.21 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr17_-_56492989 | 1.15 |
ENST00000583753.1
|
RNF43
|
ring finger protein 43 |
| chr5_+_112227311 | 1.15 |
ENST00000391338.1
|
ZRSR1
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 1 |
| chr14_-_107049312 | 1.14 |
ENST00000390627.2
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr5_-_58295712 | 1.10 |
ENST00000317118.8
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr6_-_146057144 | 1.09 |
ENST00000367519.3
|
EPM2A
|
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr2_+_29336855 | 1.07 |
ENST00000404424.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr4_+_154622652 | 1.06 |
ENST00000260010.6
|
TLR2
|
toll-like receptor 2 |
| chr18_+_61575200 | 1.04 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr7_-_115670804 | 1.02 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr12_-_51611477 | 1.01 |
ENST00000389243.4
|
POU6F1
|
POU class 6 homeobox 1 |
| chrX_-_107975917 | 1.00 |
ENST00000563887.1
|
RP6-24A23.6
|
Uncharacterized protein |
| chr13_-_46756351 | 1.00 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr2_+_29320571 | 0.98 |
ENST00000401605.1
ENST00000401617.2 |
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr7_+_150725510 | 0.98 |
ENST00000461373.1
ENST00000358849.4 ENST00000297504.6 ENST00000542328.1 ENST00000498578.1 ENST00000356058.4 ENST00000477719.1 ENST00000477092.1 |
ABCB8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr10_+_135207623 | 0.97 |
ENST00000317502.6
ENST00000432508.3 |
MTG1
|
mitochondrial ribosome-associated GTPase 1 |
| chr19_+_16940198 | 0.93 |
ENST00000248054.5
ENST00000596802.1 ENST00000379803.1 |
SIN3B
|
SIN3 transcription regulator family member B |
| chr6_-_136847610 | 0.92 |
ENST00000454590.1
ENST00000432797.2 |
MAP7
|
microtubule-associated protein 7 |
| chr10_-_112678692 | 0.90 |
ENST00000605742.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr2_-_70475730 | 0.87 |
ENST00000445587.1
ENST00000433529.2 ENST00000415783.2 |
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr14_+_21249200 | 0.85 |
ENST00000304677.2
|
RNASE6
|
ribonuclease, RNase A family, k6 |
| chr10_-_6104253 | 0.84 |
ENST00000256876.6
ENST00000379954.1 ENST00000379959.3 |
IL2RA
|
interleukin 2 receptor, alpha |
| chr16_+_31366536 | 0.83 |
ENST00000562522.1
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr12_-_23737534 | 0.82 |
ENST00000396007.2
|
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr20_+_30063067 | 0.80 |
ENST00000201979.2
|
REM1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr10_-_112678904 | 0.77 |
ENST00000423273.1
ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1
|
BBSome interacting protein 1 |
| chr3_+_70048881 | 0.76 |
ENST00000483525.1
|
RP11-460N16.1
|
RP11-460N16.1 |
| chr5_+_140501581 | 0.75 |
ENST00000194152.1
|
PCDHB4
|
protocadherin beta 4 |
| chrX_+_65382381 | 0.71 |
ENST00000519389.1
|
HEPH
|
hephaestin |
| chr10_-_115904361 | 0.71 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr8_-_110986918 | 0.67 |
ENST00000297404.1
|
KCNV1
|
potassium channel, subfamily V, member 1 |
| chr10_-_11574274 | 0.66 |
ENST00000277575.5
|
USP6NL
|
USP6 N-terminal like |
| chr4_-_68749699 | 0.65 |
ENST00000545541.1
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr6_-_27880174 | 0.64 |
ENST00000303324.2
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2 |
| chr5_-_39270725 | 0.64 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr1_+_73771844 | 0.64 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
| chr16_+_31366455 | 0.64 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr8_-_125577940 | 0.64 |
ENST00000519168.1
ENST00000395508.2 |
MTSS1
|
metastasis suppressor 1 |
| chr20_+_57875758 | 0.59 |
ENST00000395654.3
|
EDN3
|
endothelin 3 |
| chr6_-_127840453 | 0.59 |
ENST00000556132.1
|
SOGA3
|
SOGA family member 3 |
| chr1_+_22979474 | 0.58 |
ENST00000509305.1
|
C1QB
|
complement component 1, q subcomponent, B chain |
| chr15_+_58724184 | 0.57 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr14_+_31494841 | 0.55 |
ENST00000556232.1
ENST00000216366.4 ENST00000334725.4 ENST00000554609.1 ENST00000554345.1 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr5_-_59481406 | 0.53 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr12_+_54366894 | 0.53 |
ENST00000546378.1
ENST00000243082.4 |
HOXC11
|
homeobox C11 |
| chr14_-_106668095 | 0.50 |
ENST00000390606.2
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr4_-_122085469 | 0.49 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr11_-_35441597 | 0.48 |
ENST00000395753.1
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr5_-_88119580 | 0.47 |
ENST00000539796.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr8_+_70404996 | 0.47 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr10_-_22292613 | 0.46 |
ENST00000376980.3
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr17_-_40540586 | 0.45 |
ENST00000264657.5
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr10_-_112678976 | 0.43 |
ENST00000448814.1
|
BBIP1
|
BBSome interacting protein 1 |
| chrX_-_15288154 | 0.42 |
ENST00000380483.3
ENST00000380485.3 ENST00000380488.4 |
ASB9
|
ankyrin repeat and SOCS box containing 9 |
| chr15_+_25068773 | 0.41 |
ENST00000400100.1
ENST00000400098.1 |
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chr16_+_619931 | 0.41 |
ENST00000321878.5
ENST00000439574.1 ENST00000026218.5 ENST00000470411.2 |
PIGQ
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr5_+_140767452 | 0.40 |
ENST00000519479.1
|
PCDHGB4
|
protocadherin gamma subfamily B, 4 |
| chr18_-_31803435 | 0.39 |
ENST00000589544.1
ENST00000269185.4 ENST00000261592.5 |
NOL4
|
nucleolar protein 4 |
| chr2_+_169757750 | 0.39 |
ENST00000375363.3
ENST00000429379.2 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase, catalytic, 2 |
| chr16_-_11363178 | 0.35 |
ENST00000312693.3
|
TNP2
|
transition protein 2 (during histone to protamine replacement) |
| chr1_+_63063152 | 0.35 |
ENST00000371129.3
|
ANGPTL3
|
angiopoietin-like 3 |
| chr6_-_76782371 | 0.33 |
ENST00000369950.3
ENST00000369963.3 |
IMPG1
|
interphotoreceptor matrix proteoglycan 1 |
| chr14_-_65409502 | 0.32 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr2_+_202047843 | 0.28 |
ENST00000272879.5
ENST00000374650.3 ENST00000346817.5 ENST00000313728.7 ENST00000448480.1 |
CASP10
|
caspase 10, apoptosis-related cysteine peptidase |
| chr11_-_8290263 | 0.26 |
ENST00000428101.2
|
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr14_+_39703112 | 0.23 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr11_+_86085778 | 0.23 |
ENST00000354755.1
ENST00000278487.3 ENST00000531271.1 ENST00000445632.2 |
CCDC81
|
coiled-coil domain containing 81 |
| chr12_+_51985001 | 0.21 |
ENST00000354534.6
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr11_-_35441524 | 0.20 |
ENST00000395750.1
ENST00000449068.1 |
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr3_-_169587621 | 0.19 |
ENST00000523069.1
ENST00000316428.5 ENST00000264676.5 |
LRRC31
|
leucine rich repeat containing 31 |
| chr14_+_31494672 | 0.18 |
ENST00000542754.2
ENST00000313566.6 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr5_-_11589131 | 0.18 |
ENST00000511377.1
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr17_+_7155819 | 0.17 |
ENST00000570322.1
ENST00000576496.1 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr7_-_20256965 | 0.16 |
ENST00000400331.5
ENST00000332878.4 |
MACC1
|
metastasis associated in colon cancer 1 |
| chr3_-_185826855 | 0.12 |
ENST00000306376.5
|
ETV5
|
ets variant 5 |
| chr4_+_68424434 | 0.10 |
ENST00000265404.2
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr5_-_77844974 | 0.10 |
ENST00000515007.2
|
LHFPL2
|
lipoma HMGIC fusion partner-like 2 |
| chr12_+_26164645 | 0.10 |
ENST00000542004.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr9_+_35673853 | 0.09 |
ENST00000378357.4
|
CA9
|
carbonic anhydrase IX |
| chr20_-_23731893 | 0.09 |
ENST00000398402.1
|
CST1
|
cystatin SN |
| chr7_-_107880508 | 0.07 |
ENST00000425651.2
|
NRCAM
|
neuronal cell adhesion molecule |
| chr5_-_132200477 | 0.04 |
ENST00000296875.2
|
GDF9
|
growth differentiation factor 9 |
| chr19_-_23941639 | 0.02 |
ENST00000395385.3
ENST00000531570.1 ENST00000528059.1 |
ZNF681
|
zinc finger protein 681 |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU4F1_POU4F3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 14.9 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 2.5 | 29.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.7 | 6.8 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 1.2 | 2.3 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 1.1 | 3.4 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 1.1 | 31.9 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 1.0 | 3.1 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 1.0 | 5.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 1.0 | 4.0 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.8 | 2.4 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.7 | 3.7 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.7 | 3.4 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.6 | 2.6 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.6 | 1.8 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.6 | 10.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.5 | 2.6 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.5 | 1.5 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.5 | 2.3 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) bone marrow development(GO:0048539) |
| 0.4 | 4.9 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.4 | 3.7 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.3 | 3.7 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.3 | 1.8 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.3 | 4.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.3 | 1.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.3 | 4.9 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.3 | 1.4 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.3 | 1.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.3 | 0.8 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 1.5 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.2 | 2.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.2 | 2.2 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.2 | 2.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 10.4 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 0.6 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 10.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 2.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 3.0 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.8 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 4.1 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 0.6 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 1.6 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 1.3 | GO:0007512 | adult heart development(GO:0007512) |
| 0.1 | 0.7 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 3.8 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.1 | 0.9 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 1.3 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.1 | 3.9 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.1 | 0.7 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 1.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.4 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 1.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.4 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.1 | 5.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.5 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 2.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.5 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 4.8 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.6 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 6.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 3.7 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 2.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 2.1 | GO:0051384 | response to glucocorticoid(GO:0051384) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 1.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.4 | GO:0009725 | response to hormone(GO:0009725) |
| 0.0 | 0.5 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 | 10.4 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.0 | 1.6 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 5.9 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 2.4 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.0 | 3.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.2 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.0 | 0.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.8 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 1.4 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 1.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 1.5 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.3 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 2.6 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.7 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 29.8 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.6 | 4.9 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.5 | 4.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.5 | 14.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.4 | 6.8 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.4 | 1.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.3 | 0.9 | GO:0000806 | Y chromosome(GO:0000806) |
| 0.3 | 2.6 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.2 | 2.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.2 | 1.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.2 | 1.6 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.2 | 2.6 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 1.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.9 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 0.6 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.1 | 6.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 1.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.6 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 1.0 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 1.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 2.4 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 2.9 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 2.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 2.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 8.4 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 5.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.2 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 5.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 2.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 3.7 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 3.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 1.0 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 3.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 1.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 5.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 7.0 | GO:0009986 | cell surface(GO:0009986) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 10.4 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 1.4 | 10.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.8 | 2.4 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.7 | 2.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.6 | 8.0 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.5 | 1.5 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.4 | 1.8 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.4 | 29.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.4 | 4.0 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.3 | 3.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.2 | 4.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 1.7 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 1.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.2 | 0.8 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.2 | 1.6 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 3.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 3.8 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 1.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.6 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 3.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 4.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 6.8 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 3.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.4 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 2.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 3.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 2.6 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 2.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 2.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.6 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 3.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 6.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 2.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 3.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 5.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 11.3 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.1 | 2.3 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 1.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 2.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 14.8 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.6 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 1.7 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 1.5 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 1.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.7 | GO:0016248 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 3.8 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.9 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 3.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 2.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 9.5 | GO:0008270 | zinc ion binding(GO:0008270) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 30.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 5.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 4.0 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 2.6 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 6.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 11.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 3.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 4.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 3.7 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 1.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.5 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 2.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 3.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 2.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 4.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.6 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 2.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 29.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.5 | 2.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.3 | 14.9 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.3 | 10.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 4.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 4.9 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.2 | 9.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 2.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 2.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 4.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.6 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 6.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 6.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 2.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 5.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 2.5 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 1.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.4 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 1.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 3.0 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.5 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |