Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
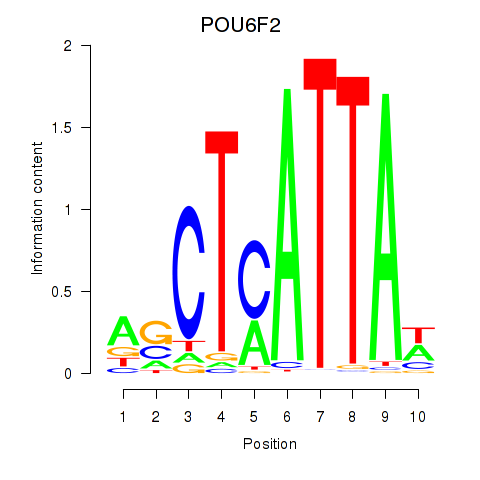
Results for POU6F2
Z-value: 0.72
Transcription factors associated with POU6F2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU6F2
|
ENSG00000106536.15 | POU class 6 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU6F2 | hg19_v2_chr7_+_39017504_39017598, hg19_v2_chr7_+_39125365_39125489 | 0.39 | 3.3e-09 | Click! |
Activity profile of POU6F2 motif
Sorted Z-values of POU6F2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_13835147 | 18.60 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr1_+_50571949 | 12.79 |
ENST00000357083.4
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr5_-_134914673 | 12.15 |
ENST00000512158.1
|
CXCL14
|
chemokine (C-X-C motif) ligand 14 |
| chr8_-_90769422 | 11.63 |
ENST00000524190.1
ENST00000523859.1 |
RP11-37B2.1
|
RP11-37B2.1 |
| chr3_+_111718036 | 10.89 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr6_+_151042224 | 10.73 |
ENST00000358517.2
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr4_+_175204818 | 10.59 |
ENST00000503780.1
|
CEP44
|
centrosomal protein 44kDa |
| chr3_+_111717600 | 10.42 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr3_+_111717511 | 10.04 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr19_-_7968427 | 9.94 |
ENST00000539278.1
|
AC010336.1
|
Uncharacterized protein |
| chr3_+_111718173 | 9.86 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr11_-_129062093 | 8.95 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr3_-_127541194 | 8.54 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase |
| chr14_+_22984601 | 8.04 |
ENST00000390509.1
|
TRAJ28
|
T cell receptor alpha joining 28 |
| chr14_-_27066960 | 7.81 |
ENST00000539517.2
|
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr17_-_29624343 | 7.43 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr12_-_16761007 | 7.35 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr1_+_10271674 | 7.05 |
ENST00000377086.1
|
KIF1B
|
kinesin family member 1B |
| chr13_+_58206655 | 6.97 |
ENST00000377918.3
|
PCDH17
|
protocadherin 17 |
| chrX_-_19988382 | 6.80 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chr22_-_50524298 | 6.77 |
ENST00000311597.5
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr3_+_189507460 | 6.72 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr3_+_159557637 | 6.71 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr22_-_50523760 | 6.58 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr1_+_50574585 | 6.23 |
ENST00000371824.1
ENST00000371823.4 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr20_+_58515417 | 6.07 |
ENST00000360816.3
|
FAM217B
|
family with sequence similarity 217, member B |
| chr7_-_73038822 | 6.04 |
ENST00000414749.2
ENST00000429400.2 ENST00000434326.1 |
MLXIPL
|
MLX interacting protein-like |
| chr13_-_110438914 | 5.89 |
ENST00000375856.3
|
IRS2
|
insulin receptor substrate 2 |
| chr5_-_140998481 | 5.71 |
ENST00000518047.1
|
DIAPH1
|
diaphanous-related formin 1 |
| chr16_-_4852915 | 5.52 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr7_-_73038867 | 5.36 |
ENST00000313375.3
ENST00000354613.1 ENST00000395189.1 ENST00000453275.1 |
MLXIPL
|
MLX interacting protein-like |
| chr12_+_54332535 | 5.31 |
ENST00000243056.3
|
HOXC13
|
homeobox C13 |
| chrX_-_38186811 | 5.23 |
ENST00000318842.7
|
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr10_+_18549645 | 4.81 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr4_-_74486347 | 4.65 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr9_-_79520989 | 4.62 |
ENST00000376713.3
ENST00000376718.3 ENST00000428286.1 |
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr11_+_45918092 | 4.26 |
ENST00000395629.2
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr3_-_57199397 | 4.24 |
ENST00000296318.7
|
IL17RD
|
interleukin 17 receptor D |
| chrX_-_38186775 | 4.14 |
ENST00000339363.3
ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr6_+_151646800 | 3.93 |
ENST00000354675.6
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr6_-_8064567 | 3.87 |
ENST00000543936.1
ENST00000397457.2 |
BLOC1S5
|
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chr14_-_50698276 | 3.58 |
ENST00000216373.5
|
SOS2
|
son of sevenless homolog 2 (Drosophila) |
| chr8_+_119294456 | 3.55 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr14_-_69261310 | 3.54 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr1_-_208417620 | 3.51 |
ENST00000367033.3
|
PLXNA2
|
plexin A2 |
| chr4_-_74486217 | 3.46 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr12_-_30887948 | 3.29 |
ENST00000433722.2
|
CAPRIN2
|
caprin family member 2 |
| chr5_+_67586465 | 3.26 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr9_+_131062367 | 3.18 |
ENST00000601297.1
|
AL359091.2
|
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr21_-_42219065 | 3.17 |
ENST00000400454.1
|
DSCAM
|
Down syndrome cell adhesion molecule |
| chr14_+_32798547 | 3.02 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr2_-_217559517 | 2.85 |
ENST00000449583.1
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr3_-_33686743 | 2.83 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr20_+_56136136 | 2.73 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr15_+_93443419 | 2.52 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chrX_+_43515467 | 2.48 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr11_-_70672645 | 2.36 |
ENST00000423696.2
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr7_-_14028488 | 2.36 |
ENST00000405358.4
|
ETV1
|
ets variant 1 |
| chr10_+_114710425 | 2.35 |
ENST00000352065.5
ENST00000369395.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr2_+_44502597 | 2.30 |
ENST00000260649.6
ENST00000409387.1 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr22_+_30792846 | 2.19 |
ENST00000312932.9
ENST00000428195.1 |
SEC14L2
|
SEC14-like 2 (S. cerevisiae) |
| chrX_+_107288239 | 2.14 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr22_-_32767017 | 2.00 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chrX_+_86772707 | 1.99 |
ENST00000373119.4
|
KLHL4
|
kelch-like family member 4 |
| chr20_+_42187608 | 1.97 |
ENST00000373100.1
|
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr4_+_88571429 | 1.95 |
ENST00000339673.6
ENST00000282479.7 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr6_+_25754927 | 1.95 |
ENST00000377905.4
ENST00000439485.2 |
SLC17A4
|
solute carrier family 17, member 4 |
| chr20_+_42187682 | 1.95 |
ENST00000373092.3
ENST00000373077.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr1_-_47655686 | 1.91 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr5_-_140998616 | 1.91 |
ENST00000389054.3
ENST00000398562.2 ENST00000389057.5 ENST00000398566.3 ENST00000398557.4 ENST00000253811.6 |
DIAPH1
|
diaphanous-related formin 1 |
| chr2_-_182545603 | 1.84 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chr6_-_111136513 | 1.77 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |
| chr14_+_32798462 | 1.75 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr6_-_133035185 | 1.74 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chrX_+_68835911 | 1.61 |
ENST00000525810.1
ENST00000527388.1 ENST00000374553.2 ENST00000374552.4 ENST00000338901.3 ENST00000524573.1 |
EDA
|
ectodysplasin A |
| chr17_-_39553844 | 1.58 |
ENST00000251645.2
|
KRT31
|
keratin 31 |
| chr11_+_65554493 | 1.57 |
ENST00000335987.3
|
OVOL1
|
ovo-like zinc finger 1 |
| chr1_-_21606013 | 1.56 |
ENST00000357071.4
|
ECE1
|
endothelin converting enzyme 1 |
| chr6_-_100912785 | 1.55 |
ENST00000369208.3
|
SIM1
|
single-minded family bHLH transcription factor 1 |
| chr7_+_129007964 | 1.48 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr2_+_138721850 | 1.48 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chrX_+_107288197 | 1.47 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr3_-_101039402 | 1.47 |
ENST00000193391.7
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr3_-_45838011 | 1.41 |
ENST00000358525.4
ENST00000413781.1 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr12_-_71031185 | 1.26 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr6_-_33160231 | 1.19 |
ENST00000395194.1
ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2
|
collagen, type XI, alpha 2 |
| chr16_+_31885079 | 1.17 |
ENST00000300870.10
ENST00000394846.3 |
ZNF267
|
zinc finger protein 267 |
| chr20_-_45035223 | 1.15 |
ENST00000450812.1
ENST00000290246.6 ENST00000439931.2 ENST00000396391.1 |
ELMO2
|
engulfment and cell motility 2 |
| chr5_-_156486120 | 1.14 |
ENST00000522693.1
|
HAVCR1
|
hepatitis A virus cellular receptor 1 |
| chr12_-_71031220 | 1.09 |
ENST00000334414.6
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr8_+_77593448 | 1.06 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr6_+_50786414 | 1.04 |
ENST00000344788.3
ENST00000393655.3 ENST00000263046.4 |
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta) |
| chr6_+_42123141 | 1.02 |
ENST00000418175.1
ENST00000541991.1 ENST00000053469.4 ENST00000394237.1 ENST00000372963.1 |
GUCA1A
RP1-139D8.6
|
guanylate cyclase activator 1A (retina) RP1-139D8.6 |
| chr8_+_77593474 | 1.02 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr3_-_18466787 | 1.00 |
ENST00000338745.6
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr17_-_40337470 | 1.00 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr15_+_58702742 | 0.99 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr7_-_14029283 | 0.95 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr12_-_53045948 | 0.87 |
ENST00000309680.3
|
KRT2
|
keratin 2 |
| chr6_-_155635583 | 0.81 |
ENST00000367166.4
|
TFB1M
|
transcription factor B1, mitochondrial |
| chr1_-_204329013 | 0.78 |
ENST00000272203.3
ENST00000414478.1 |
PLEKHA6
|
pleckstrin homology domain containing, family A member 6 |
| chr2_-_217560248 | 0.76 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chrX_-_23926004 | 0.70 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr10_-_75415825 | 0.70 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr4_+_110834033 | 0.65 |
ENST00000509793.1
ENST00000265171.5 |
EGF
|
epidermal growth factor |
| chr11_+_125034586 | 0.62 |
ENST00000298282.9
|
PKNOX2
|
PBX/knotted 1 homeobox 2 |
| chrX_+_99839799 | 0.59 |
ENST00000373031.4
|
TNMD
|
tenomodulin |
| chr3_+_189507523 | 0.57 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr12_+_15699286 | 0.57 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr10_+_120967072 | 0.52 |
ENST00000392870.2
|
GRK5
|
G protein-coupled receptor kinase 5 |
| chr6_-_89927151 | 0.44 |
ENST00000454853.2
|
GABRR1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr17_+_4613776 | 0.34 |
ENST00000269260.2
|
ARRB2
|
arrestin, beta 2 |
| chr11_+_31531291 | 0.34 |
ENST00000350638.5
ENST00000379163.5 ENST00000395934.2 |
ELP4
|
elongator acetyltransferase complex subunit 4 |
| chr19_+_50016610 | 0.25 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr2_+_169757750 | 0.14 |
ENST00000375363.3
ENST00000429379.2 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase, catalytic, 2 |
| chr1_+_175036966 | 0.11 |
ENST00000239462.4
|
TNN
|
tenascin N |
| chr9_-_13165457 | 0.08 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr12_-_86230315 | 0.01 |
ENST00000361228.3
|
RASSF9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU6F2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.1 | GO:1904647 | response to rotenone(GO:1904647) |
| 1.9 | 18.6 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.8 | 7.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 1.7 | 12.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.6 | 11.4 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 1.2 | 7.4 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 1.2 | 8.5 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 1.2 | 3.6 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 1.2 | 4.8 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 1.2 | 3.5 | GO:0048382 | mesendoderm development(GO:0048382) |
| 1.0 | 7.0 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 1.0 | 5.9 | GO:1903960 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) |
| 0.9 | 13.3 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.8 | 4.8 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.7 | 2.7 | GO:0006114 | glycerol biosynthetic process(GO:0006114) response to methionine(GO:1904640) |
| 0.6 | 7.6 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.5 | 4.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.5 | 1.6 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.5 | 5.3 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.4 | 2.8 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.4 | 1.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.4 | 1.6 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.4 | 2.3 | GO:1990822 | L-cystine transport(GO:0015811) basic amino acid transmembrane transport(GO:1990822) |
| 0.4 | 1.8 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.4 | 3.5 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.3 | 1.4 | GO:0015824 | proline transport(GO:0015824) |
| 0.3 | 3.9 | GO:0050942 | positive regulation of pigment cell differentiation(GO:0050942) |
| 0.3 | 1.6 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.3 | 3.9 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 | 1.5 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.2 | 2.0 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 3.2 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.2 | 3.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 0.6 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.2 | 3.3 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.2 | 0.6 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 1.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.2 | 1.7 | GO:2000400 | pantothenate metabolic process(GO:0015939) positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.2 | 9.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.2 | 1.0 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 2.5 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 0.6 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 1.0 | GO:0072501 | cellular divalent inorganic anion homeostasis(GO:0072501) |
| 0.1 | 3.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 2.4 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 3.3 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 2.0 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 1.0 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 1.0 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.1 | 6.5 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.1 | 1.0 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 5.5 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 11.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 1.6 | GO:0001657 | ureteric bud development(GO:0001657) mesonephros development(GO:0001823) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) |
| 0.1 | 0.5 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 3.9 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.8 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 7.9 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 0.3 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 2.2 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 4.5 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 2.5 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 15.4 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 1.8 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.0 | 1.6 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.7 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 1.2 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.7 | 4.8 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.5 | 4.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.4 | 3.6 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.4 | 1.6 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.4 | 4.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 3.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.3 | 2.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 2.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 3.9 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 41.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 13.1 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 9.4 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.1 | 1.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 7.6 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.1 | 9.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 10.6 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 8.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 2.4 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 1.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 3.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 2.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 12.1 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.0 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 1.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 3.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 7.0 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 1.1 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 3.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.0 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 2.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 4.7 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 2.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.7 | 8.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.6 | 22.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.6 | 4.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.6 | 4.8 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.6 | 7.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.5 | 2.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.5 | 8.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.4 | 2.3 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.4 | 4.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.3 | 1.7 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.3 | 2.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.3 | 3.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 3.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.3 | 2.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 5.9 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.2 | 11.7 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.2 | 1.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 3.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 1.0 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.2 | 0.5 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 41.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.2 | 1.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.2 | 2.0 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.2 | 0.8 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 3.9 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 14.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 7.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 2.9 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 2.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 13.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 0.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.9 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.0 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.6 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 1.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 2.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 7.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0031762 | alpha-1A adrenergic receptor binding(GO:0031691) follicle-stimulating hormone receptor binding(GO:0031762) platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 7.7 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 1.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.0 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 2.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 7.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.1 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 4.6 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 2.5 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 1.5 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 3.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 12.7 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 8.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 9.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 4.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 9.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 7.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 3.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 4.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 3.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 3.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 8.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 2.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.3 | 5.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 2.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 3.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 3.5 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.2 | 3.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 3.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 3.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 4.8 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 3.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 11.4 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.1 | 7.4 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.6 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 1.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 4.2 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 1.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |