Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for PPARD
Z-value: 0.56
Transcription factors associated with PPARD
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PPARD
|
ENSG00000112033.9 | peroxisome proliferator activated receptor delta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PPARD | hg19_v2_chr6_+_35310312_35310359 | -0.22 | 1.2e-03 | Click! |
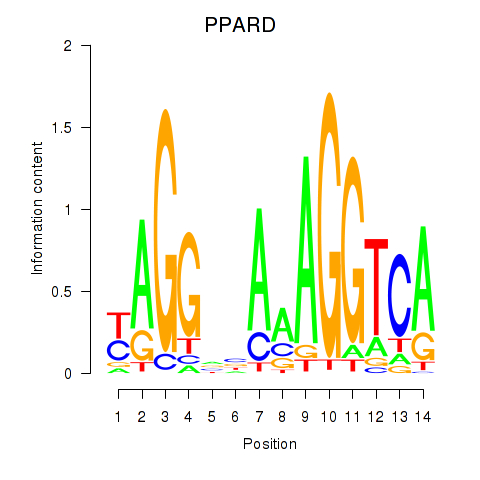
Activity profile of PPARD motif
Sorted Z-values of PPARD motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_10687948 | 5.41 |
ENST00000592285.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr2_+_120125245 | 4.80 |
ENST00000393103.2
|
DBI
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr1_+_165864821 | 4.68 |
ENST00000470820.1
|
UCK2
|
uridine-cytidine kinase 2 |
| chr12_-_53320245 | 4.60 |
ENST00000552150.1
|
KRT8
|
keratin 8 |
| chr2_+_219283815 | 4.55 |
ENST00000248444.5
ENST00000454069.1 ENST00000392114.2 |
VIL1
|
villin 1 |
| chr1_+_165864800 | 4.54 |
ENST00000469256.2
|
UCK2
|
uridine-cytidine kinase 2 |
| chr6_+_32006159 | 4.46 |
ENST00000478281.1
ENST00000471671.1 ENST00000435122.2 |
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr6_+_32006042 | 4.34 |
ENST00000418967.2
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr4_+_140586922 | 3.89 |
ENST00000265498.1
ENST00000506797.1 |
MGST2
|
microsomal glutathione S-transferase 2 |
| chr8_-_80942061 | 3.86 |
ENST00000519386.1
|
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr4_-_7069760 | 3.76 |
ENST00000264954.4
|
GRPEL1
|
GrpE-like 1, mitochondrial (E. coli) |
| chr8_-_80942139 | 3.69 |
ENST00000521434.1
ENST00000519120.1 ENST00000520946.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr11_+_66624527 | 3.68 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr17_+_37894179 | 3.66 |
ENST00000577695.1
ENST00000309156.4 ENST00000309185.3 |
GRB7
|
growth factor receptor-bound protein 7 |
| chr2_+_198365122 | 3.59 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr19_-_10687907 | 3.54 |
ENST00000589348.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr19_-_10687983 | 3.48 |
ENST00000587069.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr12_+_96252706 | 3.44 |
ENST00000266735.5
ENST00000553192.1 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr8_-_80942467 | 3.32 |
ENST00000518271.1
ENST00000276585.4 ENST00000521605.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr22_-_51016846 | 3.31 |
ENST00000312108.7
ENST00000395650.2 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr2_+_198365095 | 3.29 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr22_-_51017084 | 3.25 |
ENST00000360719.2
ENST00000457250.1 ENST00000440709.1 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr4_+_174089904 | 3.16 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr2_+_178257372 | 3.12 |
ENST00000264167.4
ENST00000409888.1 |
AGPS
|
alkylglycerone phosphate synthase |
| chr10_+_94050913 | 3.09 |
ENST00000358935.2
|
MARCH5
|
membrane-associated ring finger (C3HC4) 5 |
| chr22_+_30163340 | 2.99 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr1_-_43855444 | 2.97 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr16_-_4401258 | 2.93 |
ENST00000577031.1
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr7_-_87342564 | 2.74 |
ENST00000265724.3
ENST00000416177.1 |
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr1_-_109968973 | 2.66 |
ENST00000271308.4
ENST00000538610.1 |
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5 |
| chr2_-_207024134 | 2.58 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr1_-_120311517 | 2.51 |
ENST00000369406.3
ENST00000544913.2 |
HMGCS2
|
3-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial) |
| chr2_-_28113217 | 2.46 |
ENST00000444339.2
|
RBKS
|
ribokinase |
| chr15_-_82555000 | 2.43 |
ENST00000557844.1
ENST00000359445.3 ENST00000268206.7 |
EFTUD1
|
elongation factor Tu GTP binding domain containing 1 |
| chr17_+_7483761 | 2.35 |
ENST00000584180.1
|
CD68
|
CD68 molecule |
| chr17_+_7123125 | 2.29 |
ENST00000356839.5
ENST00000583312.1 ENST00000350303.5 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr1_-_161193349 | 2.21 |
ENST00000469730.2
ENST00000463273.1 ENST00000464492.1 ENST00000367990.3 ENST00000470459.2 ENST00000468465.1 ENST00000463812.1 |
APOA2
|
apolipoprotein A-II |
| chr19_+_17420340 | 2.14 |
ENST00000359866.4
|
DDA1
|
DET1 and DDB1 associated 1 |
| chr1_+_145727681 | 1.94 |
ENST00000417171.1
ENST00000451928.2 |
PDZK1
|
PDZ domain containing 1 |
| chr15_+_43885252 | 1.94 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr6_-_33547975 | 1.87 |
ENST00000442998.2
ENST00000360661.5 |
BAK1
|
BCL2-antagonist/killer 1 |
| chr16_-_4401284 | 1.86 |
ENST00000318059.3
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr5_+_34757309 | 1.80 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr6_-_33548006 | 1.73 |
ENST00000374467.3
|
BAK1
|
BCL2-antagonist/killer 1 |
| chr19_-_51869592 | 1.69 |
ENST00000596253.1
ENST00000309244.4 |
ETFB
|
electron-transfer-flavoprotein, beta polypeptide |
| chr11_+_64073699 | 1.63 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr9_-_33402506 | 1.55 |
ENST00000377425.4
ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7
|
aquaporin 7 |
| chr1_+_55107449 | 1.55 |
ENST00000421030.2
ENST00000545244.1 ENST00000339553.5 ENST00000409996.1 ENST00000454855.2 |
MROH7
|
maestro heat-like repeat family member 7 |
| chr15_+_43985084 | 1.52 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr6_+_33172407 | 1.50 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr5_-_143550241 | 1.41 |
ENST00000522203.1
|
YIPF5
|
Yip1 domain family, member 5 |
| chrX_-_108868390 | 1.38 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr5_-_143550159 | 1.36 |
ENST00000448443.2
ENST00000513112.1 ENST00000519064.1 ENST00000274496.5 |
YIPF5
|
Yip1 domain family, member 5 |
| chr19_+_49867181 | 1.33 |
ENST00000597546.1
|
DKKL1
|
dickkopf-like 1 |
| chr17_-_39728303 | 1.30 |
ENST00000588431.1
ENST00000246662.4 |
KRT9
|
keratin 9 |
| chrX_+_2746850 | 1.27 |
ENST00000381163.3
ENST00000338623.5 ENST00000542787.1 |
GYG2
|
glycogenin 2 |
| chr17_-_34270668 | 1.26 |
ENST00000293274.4
|
LYZL6
|
lysozyme-like 6 |
| chr17_-_34270697 | 1.25 |
ENST00000585556.1
|
LYZL6
|
lysozyme-like 6 |
| chr7_+_7222233 | 1.20 |
ENST00000436587.2
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr6_+_43266063 | 1.13 |
ENST00000372574.3
|
SLC22A7
|
solute carrier family 22 (organic anion transporter), member 7 |
| chrX_+_2746818 | 1.12 |
ENST00000398806.3
|
GYG2
|
glycogenin 2 |
| chr6_+_88299833 | 1.11 |
ENST00000392844.3
ENST00000257789.4 ENST00000546266.1 ENST00000417380.2 |
ORC3
|
origin recognition complex, subunit 3 |
| chr19_+_10196981 | 0.95 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr9_+_115142217 | 0.88 |
ENST00000398805.3
ENST00000398803.1 ENST00000262542.7 ENST00000539114.1 |
HSDL2
|
hydroxysteroid dehydrogenase like 2 |
| chr17_-_34344991 | 0.85 |
ENST00000591423.1
|
CCL23
|
chemokine (C-C motif) ligand 23 |
| chr22_-_42526802 | 0.85 |
ENST00000359033.4
ENST00000389970.3 ENST00000360608.5 |
CYP2D6
|
cytochrome P450, family 2, subfamily D, polypeptide 6 |
| chr1_-_161102367 | 0.84 |
ENST00000464113.1
|
DEDD
|
death effector domain containing |
| chr21_-_27107198 | 0.81 |
ENST00000400094.1
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr6_+_43265992 | 0.78 |
ENST00000449231.1
ENST00000372589.3 ENST00000372585.5 |
SLC22A7
|
solute carrier family 22 (organic anion transporter), member 7 |
| chr21_-_27107344 | 0.75 |
ENST00000457143.2
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr15_+_75494214 | 0.73 |
ENST00000394987.4
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr21_-_27107283 | 0.72 |
ENST00000284971.3
ENST00000400099.1 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr6_+_143999072 | 0.72 |
ENST00000440869.2
ENST00000367582.3 ENST00000451827.2 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr1_-_11863171 | 0.70 |
ENST00000376592.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr20_+_42187608 | 0.69 |
ENST00000373100.1
|
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr20_+_42187682 | 0.62 |
ENST00000373092.3
ENST00000373077.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr3_+_186383741 | 0.61 |
ENST00000232003.4
|
HRG
|
histidine-rich glycoprotein |
| chr13_+_113760098 | 0.59 |
ENST00000346342.3
ENST00000541084.1 ENST00000375581.3 |
F7
|
coagulation factor VII (serum prothrombin conversion accelerator) |
| chr21_-_27107881 | 0.58 |
ENST00000400090.3
ENST00000400087.3 ENST00000400093.3 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr18_-_47340297 | 0.58 |
ENST00000586485.1
ENST00000587994.1 ENST00000586100.1 ENST00000285093.10 |
ACAA2
|
acetyl-CoA acyltransferase 2 |
| chr15_+_78441663 | 0.57 |
ENST00000299518.2
ENST00000558554.1 ENST00000557826.1 ENST00000561279.1 ENST00000559186.1 ENST00000560770.1 ENST00000559881.1 ENST00000559205.1 ENST00000441490.2 |
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr2_-_178257401 | 0.53 |
ENST00000464747.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr17_-_34345002 | 0.52 |
ENST00000293280.2
|
CCL23
|
chemokine (C-C motif) ligand 23 |
| chr1_-_161102421 | 0.51 |
ENST00000490843.2
ENST00000368006.3 ENST00000392188.1 ENST00000545495.1 |
DEDD
|
death effector domain containing |
| chr1_+_62417957 | 0.46 |
ENST00000307297.7
ENST00000543708.1 |
INADL
|
InaD-like (Drosophila) |
| chr2_+_28113583 | 0.41 |
ENST00000344773.2
ENST00000379624.1 ENST00000342045.2 ENST00000379632.2 ENST00000361704.2 |
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator) |
| chr3_-_187009798 | 0.41 |
ENST00000337774.5
|
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
| chr11_-_119599794 | 0.30 |
ENST00000264025.3
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chr1_-_160832642 | 0.30 |
ENST00000368034.4
|
CD244
|
CD244 molecule, natural killer cell receptor 2B4 |
| chr12_-_21757774 | 0.28 |
ENST00000261195.2
|
GYS2
|
glycogen synthase 2 (liver) |
| chr20_+_46130601 | 0.28 |
ENST00000341724.6
|
NCOA3
|
nuclear receptor coactivator 3 |
| chr1_-_11107280 | 0.27 |
ENST00000400897.3
ENST00000400898.3 |
MASP2
|
mannan-binding lectin serine peptidase 2 |
| chr9_+_34458771 | 0.26 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr14_+_32546274 | 0.26 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr1_-_43855479 | 0.26 |
ENST00000290663.6
ENST00000372457.4 |
MED8
|
mediator complex subunit 8 |
| chr2_+_206547215 | 0.26 |
ENST00000360409.3
ENST00000540178.1 ENST00000540841.1 ENST00000355117.4 ENST00000450507.1 ENST00000417189.1 |
NRP2
|
neuropilin 2 |
| chr1_-_197036364 | 0.25 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr16_+_2255841 | 0.23 |
ENST00000301725.7
|
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chrX_+_38211777 | 0.23 |
ENST00000039007.4
|
OTC
|
ornithine carbamoyltransferase |
| chr16_+_2255710 | 0.21 |
ENST00000397124.1
ENST00000565250.1 |
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr11_-_47470591 | 0.14 |
ENST00000524487.1
|
RAPSN
|
receptor-associated protein of the synapse |
| chr17_+_73997419 | 0.13 |
ENST00000425876.2
|
CDK3
|
cyclin-dependent kinase 3 |
| chr2_+_73441350 | 0.09 |
ENST00000389501.4
|
SMYD5
|
SMYD family member 5 |
| chr6_+_72922590 | 0.09 |
ENST00000523963.1
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr21_+_27107672 | 0.09 |
ENST00000400075.3
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa |
| chr12_-_95467267 | 0.08 |
ENST00000330677.7
|
NR2C1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr6_+_72922505 | 0.08 |
ENST00000401910.3
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr12_+_121163538 | 0.05 |
ENST00000242592.4
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr3_-_125094093 | 0.04 |
ENST00000484491.1
ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148
|
zinc finger protein 148 |
| chr11_-_47470703 | 0.04 |
ENST00000298854.2
|
RAPSN
|
receptor-associated protein of the synapse |
| chr14_-_25045446 | 0.01 |
ENST00000216336.2
|
CTSG
|
cathepsin G |
| chr11_-_47470682 | 0.00 |
ENST00000529341.1
ENST00000352508.3 |
RAPSN
|
receptor-associated protein of the synapse |
Network of associatons between targets according to the STRING database.
First level regulatory network of PPARD
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.2 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 1.2 | 3.6 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 1.0 | 3.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.9 | 4.5 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.8 | 2.5 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.7 | 2.2 | GO:0060620 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 0.6 | 8.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.5 | 8.8 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.5 | 2.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.4 | 2.7 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.4 | 3.0 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.4 | 1.1 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.3 | 4.6 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.3 | 7.0 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.3 | 4.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.3 | 0.6 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.3 | 2.3 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.2 | 11.4 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.2 | 1.6 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.2 | 3.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 0.8 | GO:0033076 | alkaloid catabolic process(GO:0009822) isoquinoline alkaloid metabolic process(GO:0033076) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.2 | 2.5 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.2 | 2.8 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.2 | 3.1 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.2 | 3.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.2 | 3.4 | GO:0019184 | glutathione biosynthetic process(GO:0006750) nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.2 | 0.6 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 10.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.1 | 0.7 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 | 1.5 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 3.4 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.1 | 0.3 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.1 | 0.6 | GO:1902109 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 | 2.9 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 1.2 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 0.2 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.1 | 2.4 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.1 | 0.2 | GO:1900073 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.1 | 3.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 1.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 1.5 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 1.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.3 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.0 | 2.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 1.6 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.3 | GO:0097491 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.0 | 2.7 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 1.4 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 1.3 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.6 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.1 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 1.3 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 3.3 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 2.1 | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032434) |
| 0.0 | 1.4 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.5 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.5 | 2.6 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.4 | 3.4 | GO:0005683 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.4 | 10.9 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.4 | 1.1 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.3 | 3.6 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.3 | 3.0 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 11.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 4.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 2.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 2.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 2.9 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 4.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 2.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 2.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 3.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.9 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 3.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.3 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 9.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.1 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.6 | GO:0036019 | endolysosome(GO:0036019) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 2.3 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 7.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.8 | GO:0001650 | fibrillar center(GO:0001650) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 9.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 1.3 | 6.6 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 1.3 | 3.9 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.6 | 2.4 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.6 | 2.3 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.6 | 4.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.6 | 2.2 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.5 | 2.7 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.5 | 2.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.5 | 1.9 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.5 | 4.8 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.4 | 3.0 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.4 | 2.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.4 | 1.6 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.3 | 3.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.3 | 1.5 | GO:0070404 | NADH binding(GO:0070404) |
| 0.3 | 9.6 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 3.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 1.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.2 | 1.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 3.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.6 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.7 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 2.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 3.1 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 2.7 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.2 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 2.5 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 0.3 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.1 | 1.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.6 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.1 | 1.9 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.1 | 1.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 2.7 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 3.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 3.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 3.2 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 2.9 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 1.6 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.5 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 8.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 2.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.4 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 11.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.4 | 8.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.3 | 9.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 3.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 2.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 2.9 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 5.0 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 8.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 2.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 3.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 3.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 4.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 2.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 2.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 3.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 2.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 7.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 0.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.8 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 2.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 3.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |