Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
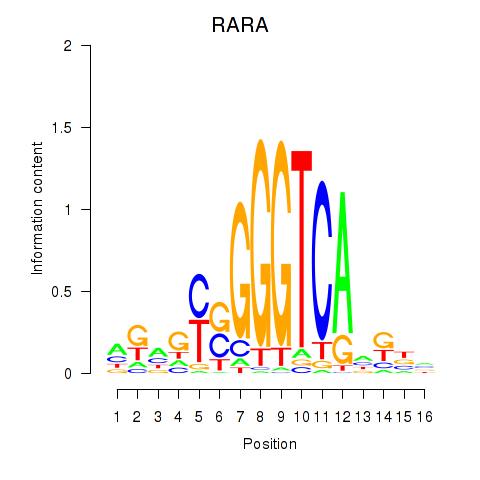
Results for RARA
Z-value: 0.74
Transcription factors associated with RARA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RARA
|
ENSG00000131759.13 | retinoic acid receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RARA | hg19_v2_chr17_+_38474489_38474548 | -0.04 | 6.0e-01 | Click! |
Activity profile of RARA motif
Sorted Z-values of RARA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_54378923 | 8.48 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr4_+_107236692 | 7.31 |
ENST00000510207.1
|
AIMP1
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chr13_+_43597269 | 7.21 |
ENST00000379221.2
|
DNAJC15
|
DnaJ (Hsp40) homolog, subfamily C, member 15 |
| chr16_-_11680791 | 6.37 |
ENST00000571976.1
ENST00000413364.2 |
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr10_+_17270214 | 6.16 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr10_+_115438920 | 5.72 |
ENST00000429617.1
ENST00000369331.4 |
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr5_+_65440032 | 5.47 |
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr1_-_53704157 | 4.71 |
ENST00000371466.4
ENST00000371470.3 |
MAGOH
|
mago-nashi homolog, proliferation-associated (Drosophila) |
| chr16_-_11680759 | 4.64 |
ENST00000571459.1
ENST00000570798.1 ENST00000572255.1 ENST00000574763.1 ENST00000574703.1 ENST00000571277.1 ENST00000381810.3 |
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr4_+_107236847 | 4.52 |
ENST00000358008.3
|
AIMP1
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chr4_+_107236722 | 4.42 |
ENST00000442366.1
|
AIMP1
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chr11_-_102323489 | 4.28 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr19_+_16435625 | 4.23 |
ENST00000248071.5
ENST00000592003.1 |
KLF2
|
Kruppel-like factor 2 |
| chr1_+_165796753 | 4.21 |
ENST00000367879.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr3_+_133293278 | 4.16 |
ENST00000508481.1
ENST00000420115.2 ENST00000504867.1 ENST00000507408.1 ENST00000511392.1 ENST00000515421.1 |
CDV3
|
CDV3 homolog (mouse) |
| chr2_-_151344172 | 4.10 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr22_+_45072925 | 4.09 |
ENST00000006251.7
|
PRR5
|
proline rich 5 (renal) |
| chr1_-_3816779 | 4.06 |
ENST00000361605.3
|
C1orf174
|
chromosome 1 open reading frame 174 |
| chr22_+_45072958 | 4.01 |
ENST00000403581.1
|
PRR5
|
proline rich 5 (renal) |
| chr9_-_21995300 | 3.98 |
ENST00000498628.2
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr3_-_185641681 | 3.97 |
ENST00000259043.7
|
TRA2B
|
transformer 2 beta homolog (Drosophila) |
| chr7_-_45960850 | 3.79 |
ENST00000381083.4
ENST00000381086.5 ENST00000275521.6 |
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr1_-_16939976 | 3.73 |
ENST00000430580.2
|
NBPF1
|
neuroblastoma breakpoint family, member 1 |
| chr11_+_32605350 | 3.73 |
ENST00000531120.1
ENST00000524896.1 ENST00000323213.5 |
EIF3M
|
eukaryotic translation initiation factor 3, subunit M |
| chr12_+_104324112 | 3.69 |
ENST00000299767.5
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr6_-_74233480 | 3.67 |
ENST00000455918.1
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr10_+_43633914 | 3.66 |
ENST00000374466.3
ENST00000374464.1 |
CSGALNACT2
|
chondroitin sulfate N-acetylgalactosaminyltransferase 2 |
| chr6_-_35888905 | 3.52 |
ENST00000510290.1
ENST00000423325.2 ENST00000373822.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr3_-_55521323 | 3.49 |
ENST00000264634.4
|
WNT5A
|
wingless-type MMTV integration site family, member 5A |
| chr15_+_90777424 | 3.43 |
ENST00000561433.1
ENST00000559204.1 ENST00000558291.1 |
GDPGP1
|
GDP-D-glucose phosphorylase 1 |
| chr9_-_21994597 | 3.42 |
ENST00000579755.1
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr9_-_21994344 | 3.38 |
ENST00000530628.2
ENST00000361570.3 |
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr1_+_145439306 | 3.37 |
ENST00000425134.1
|
TXNIP
|
thioredoxin interacting protein |
| chr11_+_114310237 | 3.36 |
ENST00000539119.1
|
REXO2
|
RNA exonuclease 2 |
| chr7_-_105752971 | 3.22 |
ENST00000011473.2
|
SYPL1
|
synaptophysin-like 1 |
| chr3_+_133292574 | 3.15 |
ENST00000264993.3
|
CDV3
|
CDV3 homolog (mouse) |
| chr1_-_93426998 | 3.14 |
ENST00000370310.4
|
FAM69A
|
family with sequence similarity 69, member A |
| chr22_+_50624323 | 3.12 |
ENST00000380909.4
ENST00000303434.4 |
TRABD
|
TraB domain containing |
| chr19_-_4540486 | 3.08 |
ENST00000306390.6
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr11_+_9482551 | 3.08 |
ENST00000438144.2
ENST00000526657.1 ENST00000299606.2 ENST00000534265.1 ENST00000412390.2 |
ZNF143
|
zinc finger protein 143 |
| chr11_+_114310164 | 3.01 |
ENST00000544196.1
ENST00000539754.1 ENST00000539275.1 |
REXO2
|
RNA exonuclease 2 |
| chr4_-_87855851 | 2.98 |
ENST00000473559.1
|
C4orf36
|
chromosome 4 open reading frame 36 |
| chr13_+_25670268 | 2.97 |
ENST00000281589.3
|
PABPC3
|
poly(A) binding protein, cytoplasmic 3 |
| chr7_+_148395733 | 2.91 |
ENST00000602748.1
|
CUL1
|
cullin 1 |
| chr1_+_25870070 | 2.89 |
ENST00000374338.4
|
LDLRAP1
|
low density lipoprotein receptor adaptor protein 1 |
| chr20_-_34330129 | 2.89 |
ENST00000397370.3
ENST00000528062.3 ENST00000407261.4 ENST00000374038.3 ENST00000361162.6 |
RBM39
|
RNA binding motif protein 39 |
| chr5_-_114961858 | 2.88 |
ENST00000282382.4
ENST00000456936.3 ENST00000408996.4 |
TMED7-TICAM2
TMED7
TICAM2
|
TMED7-TICAM2 readthrough transmembrane emp24 protein transport domain containing 7 toll-like receptor adaptor molecule 2 |
| chr6_-_31324943 | 2.82 |
ENST00000412585.2
ENST00000434333.1 |
HLA-B
|
major histocompatibility complex, class I, B |
| chr18_-_812231 | 2.80 |
ENST00000314574.4
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr5_+_118690466 | 2.75 |
ENST00000503646.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr1_+_203764742 | 2.74 |
ENST00000432282.1
ENST00000453771.1 ENST00000367214.1 ENST00000367212.3 ENST00000332127.4 |
ZC3H11A
|
zinc finger CCCH-type containing 11A |
| chr7_-_105752651 | 2.69 |
ENST00000470347.1
ENST00000455385.2 |
SYPL1
|
synaptophysin-like 1 |
| chr11_+_58910201 | 2.65 |
ENST00000528737.1
|
FAM111A
|
family with sequence similarity 111, member A |
| chr11_-_46142615 | 2.62 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr17_+_7476136 | 2.58 |
ENST00000582169.1
ENST00000578754.1 ENST00000578495.1 ENST00000293831.8 ENST00000380512.5 ENST00000585024.1 ENST00000583802.1 ENST00000577269.1 ENST00000584784.1 ENST00000582746.1 |
EIF4A1
|
eukaryotic translation initiation factor 4A1 |
| chr15_+_23255242 | 2.53 |
ENST00000450802.3
|
GOLGA8I
|
golgin A8 family, member I |
| chr7_+_148395959 | 2.46 |
ENST00000325222.4
|
CUL1
|
cullin 1 |
| chr19_-_12405689 | 2.45 |
ENST00000355684.5
|
ZNF44
|
zinc finger protein 44 |
| chr22_-_29138386 | 2.43 |
ENST00000544772.1
|
CHEK2
|
checkpoint kinase 2 |
| chr7_+_130794846 | 2.35 |
ENST00000421797.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr18_+_12948000 | 2.32 |
ENST00000585730.1
ENST00000399892.2 ENST00000589446.1 ENST00000587761.1 |
SEH1L
|
SEH1-like (S. cerevisiae) |
| chr9_-_88969303 | 2.30 |
ENST00000277141.6
ENST00000375963.3 |
ZCCHC6
|
zinc finger, CCHC domain containing 6 |
| chr5_-_157286104 | 2.29 |
ENST00000530742.1
ENST00000523908.1 ENST00000523094.1 ENST00000296951.5 ENST00000411809.2 |
CLINT1
|
clathrin interactor 1 |
| chr10_+_60144782 | 2.29 |
ENST00000487519.1
|
TFAM
|
transcription factor A, mitochondrial |
| chr15_+_80351977 | 2.29 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr11_+_10326612 | 2.29 |
ENST00000534464.1
ENST00000530439.1 ENST00000524948.1 ENST00000528655.1 ENST00000526492.1 ENST00000525063.1 |
ADM
|
adrenomedullin |
| chr16_-_425205 | 2.27 |
ENST00000448854.1
|
TMEM8A
|
transmembrane protein 8A |
| chr5_+_138609441 | 2.24 |
ENST00000509990.1
ENST00000506147.1 ENST00000512107.1 |
MATR3
|
matrin 3 |
| chr11_+_101983176 | 2.22 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chrX_+_69509927 | 2.21 |
ENST00000374403.3
|
KIF4A
|
kinesin family member 4A |
| chr22_+_24951436 | 2.19 |
ENST00000215829.3
|
SNRPD3
|
small nuclear ribonucleoprotein D3 polypeptide 18kDa |
| chr3_-_142608001 | 2.18 |
ENST00000295992.3
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr22_-_32058166 | 2.18 |
ENST00000435900.1
ENST00000336566.4 |
PISD
|
phosphatidylserine decarboxylase |
| chr15_-_63448973 | 2.17 |
ENST00000462430.1
|
RPS27L
|
ribosomal protein S27-like |
| chr2_+_189157498 | 2.15 |
ENST00000359135.3
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr9_+_114393634 | 2.13 |
ENST00000556107.1
ENST00000374294.3 |
DNAJC25
DNAJC25-GNG10
|
DnaJ (Hsp40) homolog, subfamily C , member 25 DNAJC25-GNG10 readthrough |
| chr18_-_812517 | 2.12 |
ENST00000584307.1
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr10_+_62538089 | 2.12 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr18_+_19749386 | 2.11 |
ENST00000269216.3
|
GATA6
|
GATA binding protein 6 |
| chr1_-_43855444 | 2.09 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr11_-_18610275 | 2.09 |
ENST00000543987.1
|
UEVLD
|
UEV and lactate/malate dehyrogenase domains |
| chr22_-_29137771 | 2.08 |
ENST00000439200.1
ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2
|
checkpoint kinase 2 |
| chr9_-_88969339 | 2.07 |
ENST00000375960.2
ENST00000375961.2 |
ZCCHC6
|
zinc finger, CCHC domain containing 6 |
| chr8_+_70404996 | 2.06 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr9_-_135230336 | 2.05 |
ENST00000224140.5
ENST00000372169.2 ENST00000393220.1 |
SETX
|
senataxin |
| chr14_-_71107921 | 2.04 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr8_-_101348408 | 2.02 |
ENST00000519527.1
ENST00000522369.1 |
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr10_+_60145155 | 2.02 |
ENST00000373895.3
|
TFAM
|
transcription factor A, mitochondrial |
| chr6_-_41040195 | 2.00 |
ENST00000463088.1
ENST00000469104.1 ENST00000486443.1 |
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr2_+_189157536 | 2.00 |
ENST00000409580.1
ENST00000409637.3 |
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr14_-_78227476 | 1.99 |
ENST00000554775.1
ENST00000555761.1 ENST00000554324.1 ENST00000261531.7 |
SNW1
|
SNW domain containing 1 |
| chr14_-_90085458 | 1.98 |
ENST00000345097.4
ENST00000555855.1 ENST00000555353.1 |
FOXN3
|
forkhead box N3 |
| chr14_+_24605389 | 1.98 |
ENST00000382708.3
ENST00000561435.1 |
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr13_+_53029564 | 1.98 |
ENST00000468284.1
ENST00000378034.3 ENST00000258607.5 ENST00000378037.5 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr10_-_103880209 | 1.96 |
ENST00000425280.1
|
LDB1
|
LIM domain binding 1 |
| chr1_-_33168336 | 1.95 |
ENST00000373484.3
|
SYNC
|
syncoilin, intermediate filament protein |
| chr18_+_12947981 | 1.95 |
ENST00000262124.11
|
SEH1L
|
SEH1-like (S. cerevisiae) |
| chr10_-_135090360 | 1.93 |
ENST00000486609.1
ENST00000445355.3 ENST00000485491.2 |
ADAM8
|
ADAM metallopeptidase domain 8 |
| chr11_-_18610214 | 1.93 |
ENST00000300038.7
ENST00000396197.3 ENST00000320750.6 |
UEVLD
|
UEV and lactate/malate dehyrogenase domains |
| chr3_-_142607740 | 1.92 |
ENST00000485766.1
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr5_+_31532373 | 1.92 |
ENST00000325366.9
ENST00000355907.3 ENST00000507818.2 |
C5orf22
|
chromosome 5 open reading frame 22 |
| chr12_+_51632638 | 1.91 |
ENST00000549732.2
|
DAZAP2
|
DAZ associated protein 2 |
| chr21_-_46707793 | 1.88 |
ENST00000331343.7
ENST00000349485.5 |
POFUT2
|
protein O-fucosyltransferase 2 |
| chr10_+_70715884 | 1.87 |
ENST00000354185.4
|
DDX21
|
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr14_-_75536182 | 1.85 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr21_+_35445827 | 1.84 |
ENST00000608209.1
ENST00000381151.3 |
SLC5A3
SLC5A3
|
sodium/myo-inositol cotransporter solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
| chr9_-_139891165 | 1.84 |
ENST00000494426.1
|
CLIC3
|
chloride intracellular channel 3 |
| chr13_+_53030107 | 1.84 |
ENST00000490903.1
ENST00000480747.1 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr1_+_32687971 | 1.82 |
ENST00000373586.1
|
EIF3I
|
eukaryotic translation initiation factor 3, subunit I |
| chr1_+_109289279 | 1.81 |
ENST00000370008.3
|
STXBP3
|
syntaxin binding protein 3 |
| chr9_-_132597529 | 1.81 |
ENST00000372447.3
|
C9orf78
|
chromosome 9 open reading frame 78 |
| chr9_-_21995249 | 1.80 |
ENST00000494262.1
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr17_-_80291818 | 1.79 |
ENST00000269389.3
ENST00000581691.1 |
SECTM1
|
secreted and transmembrane 1 |
| chr10_+_62538248 | 1.79 |
ENST00000448257.2
|
CDK1
|
cyclin-dependent kinase 1 |
| chr3_+_127317066 | 1.77 |
ENST00000265056.7
|
MCM2
|
minichromosome maintenance complex component 2 |
| chr15_+_28623784 | 1.76 |
ENST00000526619.2
ENST00000337838.7 ENST00000532622.2 |
GOLGA8F
|
golgin A8 family, member F |
| chr10_-_135090338 | 1.75 |
ENST00000415217.3
|
ADAM8
|
ADAM metallopeptidase domain 8 |
| chr17_+_7210898 | 1.74 |
ENST00000572815.1
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr1_-_211848899 | 1.72 |
ENST00000366998.3
ENST00000540251.1 ENST00000366999.4 |
NEK2
|
NIMA-related kinase 2 |
| chr5_-_31532160 | 1.72 |
ENST00000511367.2
ENST00000513349.1 |
DROSHA
|
drosha, ribonuclease type III |
| chr11_-_65430554 | 1.69 |
ENST00000308639.9
ENST00000406246.3 |
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr8_+_87354945 | 1.67 |
ENST00000517970.1
|
WWP1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr15_-_37393406 | 1.65 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr13_+_73356197 | 1.65 |
ENST00000326291.6
|
PIBF1
|
progesterone immunomodulatory binding factor 1 |
| chr16_+_56995762 | 1.64 |
ENST00000200676.3
ENST00000379780.2 |
CETP
|
cholesteryl ester transfer protein, plasma |
| chr1_+_26606608 | 1.64 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr6_+_111195973 | 1.61 |
ENST00000368885.3
ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr19_-_19739007 | 1.60 |
ENST00000586703.1
ENST00000591042.1 ENST00000407877.3 |
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr3_-_126194707 | 1.60 |
ENST00000336332.5
ENST00000389709.3 |
ZXDC
|
ZXD family zinc finger C |
| chr12_+_22778116 | 1.58 |
ENST00000538218.1
|
ETNK1
|
ethanolamine kinase 1 |
| chrX_-_37706815 | 1.57 |
ENST00000378578.4
|
DYNLT3
|
dynein, light chain, Tctex-type 3 |
| chr7_-_55640176 | 1.57 |
ENST00000285279.5
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr2_+_101179152 | 1.55 |
ENST00000264254.6
|
PDCL3
|
phosducin-like 3 |
| chr16_+_577697 | 1.55 |
ENST00000562370.1
ENST00000568988.1 ENST00000219611.2 |
CAPN15
|
calpain 15 |
| chr4_+_38665810 | 1.54 |
ENST00000261438.5
ENST00000514033.1 |
KLF3
|
Kruppel-like factor 3 (basic) |
| chr6_-_127664736 | 1.53 |
ENST00000368291.2
ENST00000309620.9 ENST00000454859.3 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr1_+_23037323 | 1.51 |
ENST00000544305.1
ENST00000374630.3 ENST00000400191.3 ENST00000374632.3 |
EPHB2
|
EPH receptor B2 |
| chr22_+_29664248 | 1.50 |
ENST00000406548.1
ENST00000437155.2 ENST00000415761.1 ENST00000331029.7 |
EWSR1
|
EWS RNA-binding protein 1 |
| chr1_+_76251879 | 1.50 |
ENST00000535300.1
ENST00000319942.3 |
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr5_-_137911049 | 1.49 |
ENST00000297185.3
|
HSPA9
|
heat shock 70kDa protein 9 (mortalin) |
| chr5_-_79950371 | 1.47 |
ENST00000511032.1
ENST00000504396.1 ENST00000505337.1 |
DHFR
|
dihydrofolate reductase |
| chr16_+_31885079 | 1.46 |
ENST00000300870.10
ENST00000394846.3 |
ZNF267
|
zinc finger protein 267 |
| chr14_-_91526462 | 1.45 |
ENST00000536315.2
|
RPS6KA5
|
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
| chr6_-_127664683 | 1.45 |
ENST00000528402.1
ENST00000454591.2 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr6_-_41040268 | 1.44 |
ENST00000373154.2
ENST00000244558.9 ENST00000464633.1 ENST00000424266.2 ENST00000479950.1 ENST00000482515.1 |
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr10_-_126849588 | 1.43 |
ENST00000411419.2
|
CTBP2
|
C-terminal binding protein 2 |
| chr1_-_156786530 | 1.41 |
ENST00000368198.3
|
SH2D2A
|
SH2 domain containing 2A |
| chr6_+_96025341 | 1.41 |
ENST00000369293.1
ENST00000358812.4 |
MANEA
|
mannosidase, endo-alpha |
| chr13_+_98612446 | 1.40 |
ENST00000496368.1
ENST00000421861.2 ENST00000357602.3 |
IPO5
|
importin 5 |
| chr6_+_32709119 | 1.40 |
ENST00000374940.3
|
HLA-DQA2
|
major histocompatibility complex, class II, DQ alpha 2 |
| chr22_+_50312379 | 1.37 |
ENST00000407217.3
ENST00000403427.3 |
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr1_-_113498943 | 1.35 |
ENST00000369626.3
|
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr22_+_50312316 | 1.33 |
ENST00000328268.4
|
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr2_+_47630255 | 1.33 |
ENST00000406134.1
|
MSH2
|
mutS homolog 2 |
| chr2_-_40006357 | 1.33 |
ENST00000505747.1
|
THUMPD2
|
THUMP domain containing 2 |
| chr11_-_18610246 | 1.33 |
ENST00000379387.4
ENST00000541984.1 |
UEVLD
|
UEV and lactate/malate dehyrogenase domains |
| chr1_+_215256467 | 1.32 |
ENST00000391894.2
ENST00000444842.2 |
KCNK2
|
potassium channel, subfamily K, member 2 |
| chr5_+_139781393 | 1.32 |
ENST00000360839.2
ENST00000297183.6 ENST00000421134.1 ENST00000394723.3 ENST00000511151.1 |
ANKHD1
|
ankyrin repeat and KH domain containing 1 |
| chr2_+_108443388 | 1.31 |
ENST00000354986.4
ENST00000408999.3 |
RGPD4
|
RANBP2-like and GRIP domain containing 4 |
| chr2_-_235405679 | 1.30 |
ENST00000390645.2
|
ARL4C
|
ADP-ribosylation factor-like 4C |
| chr16_-_15188106 | 1.29 |
ENST00000429751.2
ENST00000564131.1 ENST00000563559.1 ENST00000198767.6 |
RRN3
|
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) |
| chr1_-_113498616 | 1.28 |
ENST00000433570.4
ENST00000538576.1 ENST00000458229.1 |
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr11_-_8986474 | 1.28 |
ENST00000525069.1
|
TMEM9B
|
TMEM9 domain family, member B |
| chr11_-_65325430 | 1.26 |
ENST00000322147.4
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr12_+_67663056 | 1.26 |
ENST00000545606.1
|
CAND1
|
cullin-associated and neddylation-dissociated 1 |
| chr5_+_112312416 | 1.26 |
ENST00000389063.2
|
DCP2
|
decapping mRNA 2 |
| chr11_+_59480899 | 1.24 |
ENST00000300150.7
|
STX3
|
syntaxin 3 |
| chr15_+_49462434 | 1.24 |
ENST00000558145.1
ENST00000543495.1 ENST00000544523.1 ENST00000560138.1 |
GALK2
|
galactokinase 2 |
| chr5_-_146781153 | 1.24 |
ENST00000520473.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr9_+_21802542 | 1.23 |
ENST00000380172.4
|
MTAP
|
methylthioadenosine phosphorylase |
| chrX_+_100878079 | 1.23 |
ENST00000471229.2
|
ARMCX3
|
armadillo repeat containing, X-linked 3 |
| chr15_-_74374891 | 1.23 |
ENST00000290438.3
|
GOLGA6A
|
golgin A6 family, member A |
| chr15_+_33010175 | 1.22 |
ENST00000300177.4
ENST00000560677.1 ENST00000560830.1 |
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr19_-_12405606 | 1.22 |
ENST00000356109.5
|
ZNF44
|
zinc finger protein 44 |
| chr11_+_105948216 | 1.20 |
ENST00000278618.4
|
AASDHPPT
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr3_-_52486841 | 1.20 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr19_-_16770915 | 1.20 |
ENST00000593459.1
ENST00000358726.6 ENST00000597711.1 ENST00000487416.2 |
CTC-429P9.4
SMIM7
|
Small integral membrane protein 7; Uncharacterized protein small integral membrane protein 7 |
| chr10_-_112678692 | 1.19 |
ENST00000605742.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr10_-_112678904 | 1.19 |
ENST00000423273.1
ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1
|
BBSome interacting protein 1 |
| chr2_+_209130965 | 1.19 |
ENST00000392202.3
ENST00000264380.4 ENST00000407449.1 ENST00000308862.6 |
PIKFYVE
|
phosphoinositide kinase, FYVE finger containing |
| chr16_+_29674277 | 1.18 |
ENST00000395389.2
|
SPN
|
sialophorin |
| chr17_+_57784826 | 1.18 |
ENST00000262291.4
|
VMP1
|
vacuole membrane protein 1 |
| chr1_-_156786634 | 1.18 |
ENST00000392306.2
ENST00000368199.3 |
SH2D2A
|
SH2 domain containing 2A |
| chr3_+_157827841 | 1.17 |
ENST00000295930.3
ENST00000471994.1 ENST00000464171.1 ENST00000312179.6 ENST00000475278.2 |
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr1_-_111506562 | 1.17 |
ENST00000485275.2
ENST00000369763.4 |
LRIF1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr6_-_127664475 | 1.17 |
ENST00000474289.2
ENST00000534442.1 ENST00000368289.2 ENST00000525745.1 ENST00000430841.2 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr6_+_32006159 | 1.16 |
ENST00000478281.1
ENST00000471671.1 ENST00000435122.2 |
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr6_+_36097992 | 1.16 |
ENST00000211287.4
|
MAPK13
|
mitogen-activated protein kinase 13 |
| chr14_-_64194745 | 1.16 |
ENST00000247225.6
|
SGPP1
|
sphingosine-1-phosphate phosphatase 1 |
| chr22_-_31328881 | 1.15 |
ENST00000445980.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chr2_-_113594279 | 1.15 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr11_+_44117741 | 1.14 |
ENST00000395673.3
ENST00000343631.3 |
EXT2
|
exostosin glycosyltransferase 2 |
| chr5_+_140019004 | 1.14 |
ENST00000394671.3
ENST00000511410.1 ENST00000537378.1 |
TMCO6
|
transmembrane and coiled-coil domains 6 |
| chr10_-_43892668 | 1.14 |
ENST00000544000.1
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr19_+_55105085 | 1.14 |
ENST00000251372.3
ENST00000453777.1 |
LILRA1
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 1 |
| chr2_+_47630108 | 1.13 |
ENST00000233146.2
ENST00000454849.1 ENST00000543555.1 |
MSH2
|
mutS homolog 2 |
| chr12_+_22778009 | 1.12 |
ENST00000266517.4
ENST00000335148.3 |
ETNK1
|
ethanolamine kinase 1 |
| chr6_+_32006042 | 1.11 |
ENST00000418967.2
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr4_-_2936514 | 1.11 |
ENST00000508221.1
ENST00000507555.1 ENST00000355443.4 |
MFSD10
|
major facilitator superfamily domain containing 10 |
| chr14_-_105262055 | 1.08 |
ENST00000349310.3
|
AKT1
|
v-akt murine thymoma viral oncogene homolog 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RARA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 1.4 | 4.2 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 1.2 | 13.5 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 1.2 | 3.7 | GO:2000412 | activation of MAPK activity involved in innate immune response(GO:0035419) positive regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000309) positive regulation of thymocyte migration(GO:2000412) |
| 1.2 | 3.5 | GO:1904933 | cardiac right atrium morphogenesis(GO:0003213) negative regulation of melanin biosynthetic process(GO:0048022) positive regulation of anagen(GO:0051885) mediolateral intercalation(GO:0060031) cervix development(GO:0060067) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) negative regulation of secondary metabolite biosynthetic process(GO:1900377) regulation of cell proliferation in midbrain(GO:1904933) |
| 1.1 | 4.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 1.1 | 4.2 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 1.0 | 4.0 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.9 | 8.5 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.9 | 5.6 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.9 | 3.7 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.7 | 8.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.7 | 1.4 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.7 | 2.9 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.7 | 5.7 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.7 | 2.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.7 | 6.6 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.7 | 2.6 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.7 | 3.9 | GO:1902957 | negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
| 0.6 | 1.1 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.6 | 6.2 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.5 | 1.6 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.5 | 2.0 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.5 | 1.0 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.5 | 2.5 | GO:0006311 | meiotic gene conversion(GO:0006311) regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.5 | 2.0 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.5 | 2.9 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.5 | 2.3 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.4 | 1.3 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.4 | 1.2 | GO:0070638 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.4 | 1.2 | GO:1900158 | negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.4 | 3.6 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.4 | 2.0 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.4 | 1.9 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.4 | 1.5 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.4 | 1.5 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.4 | 18.3 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.4 | 3.5 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.3 | 4.9 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.3 | 1.7 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.3 | 1.0 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.3 | 5.6 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.3 | 1.3 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.3 | 3.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.3 | 0.9 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.3 | 0.6 | GO:1902613 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.3 | 1.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.3 | 1.2 | GO:0002884 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of hypersensitivity(GO:0002884) |
| 0.3 | 1.2 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.3 | 0.9 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.3 | 0.9 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.3 | 2.8 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.3 | 4.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.3 | 1.3 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.3 | 9.8 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.3 | 3.7 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.3 | 1.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.2 | 0.7 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.2 | 2.2 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.2 | 1.7 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.2 | 1.0 | GO:0048861 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.2 | 0.7 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.2 | 0.7 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.2 | 2.3 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.2 | 0.7 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.2 | 1.8 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.2 | 1.7 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 3.8 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.2 | 1.9 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.2 | 7.6 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.2 | 1.0 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) |
| 0.2 | 1.8 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.2 | 1.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 0.6 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.2 | 1.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.2 | 1.5 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 1.3 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.2 | 1.8 | GO:0015791 | polyol transport(GO:0015791) |
| 0.2 | 2.7 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.2 | 10.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 1.2 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 3.4 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.2 | 4.9 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.2 | 0.3 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.2 | 0.5 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.2 | 0.7 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.2 | 1.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 2.1 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.2 | 0.8 | GO:0061205 | alveolar primary septum development(GO:0061143) paramesonephric duct development(GO:0061205) |
| 0.2 | 1.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 0.6 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.2 | 0.9 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 1.0 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.1 | 0.6 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.1 | 2.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.7 | GO:0051586 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.1 | 0.6 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 0.1 | 2.7 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 2.2 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.1 | 1.7 | GO:0051823 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.4 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.1 | 2.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.9 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) protein localization to adherens junction(GO:0071896) |
| 0.1 | 2.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 0.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 1.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.6 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 2.4 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 2.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.8 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.1 | 3.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 3.0 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.1 | 4.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.5 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.1 | 0.5 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.7 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.6 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.5 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.5 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 0.5 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 0.6 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 0.5 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 0.8 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.1 | 0.5 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 0.9 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 1.5 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.1 | 1.6 | GO:0072662 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 0.2 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.1 | 0.5 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 2.3 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 1.0 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.1 | 1.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 2.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 2.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 1.0 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 1.3 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.2 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 1.9 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 0.9 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 1.0 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.9 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.7 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 1.7 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.0 | 0.2 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.3 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.7 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.1 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) histone H4-R3 methylation(GO:0043985) |
| 0.0 | 1.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.0 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 1.0 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 1.1 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 1.0 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.0 | 0.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 4.0 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.5 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 1.8 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.0 | 2.6 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 1.2 | GO:0006363 | transcription elongation from RNA polymerase I promoter(GO:0006362) termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 2.2 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.0 | 1.1 | GO:0015893 | drug transport(GO:0015893) |
| 0.0 | 1.1 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.7 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 2.4 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 2.1 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.0 | 0.6 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 1.2 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.7 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 1.8 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 2.7 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.7 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 3.2 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.7 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 1.4 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.6 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.7 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.6 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 2.6 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.8 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 2.3 | GO:0010952 | positive regulation of peptidase activity(GO:0010952) |
| 0.0 | 0.5 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0042699 | primary ovarian follicle growth(GO:0001545) follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.2 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 1.2 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.9 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.0 | GO:0006110 | regulation of glycolytic process(GO:0006110) |
| 0.0 | 0.2 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.6 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.2 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 1.3 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 16.3 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 1.8 | 11.0 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 1.2 | 13.5 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 1.2 | 3.7 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.9 | 8.5 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.8 | 2.5 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.8 | 3.9 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.8 | 5.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.6 | 5.6 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.6 | 3.7 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.6 | 4.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.6 | 1.7 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.5 | 3.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.5 | 8.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.4 | 2.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.4 | 3.8 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.4 | 6.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 3.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.3 | 2.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.3 | 1.2 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.3 | 0.9 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.3 | 2.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.3 | 1.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.3 | 2.2 | GO:0005683 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.3 | 2.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 1.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 4.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 0.9 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 1.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 2.0 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.2 | 2.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 0.6 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.2 | 1.8 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 2.7 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.2 | 1.2 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.2 | 1.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 1.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.2 | 0.6 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 1.3 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 1.7 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.9 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 0.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.5 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 3.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.5 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.1 | 9.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 1.0 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 5.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 2.0 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 3.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 1.3 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 1.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 0.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.4 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 1.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 4.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 16.7 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 1.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 0.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.7 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 0.3 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.1 | 1.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 1.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 1.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 2.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 1.0 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 2.3 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 7.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 2.3 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 1.0 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 3.5 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 2.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 1.6 | GO:0043234 | protein complex(GO:0043234) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 2.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 2.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 3.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 1.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 2.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 1.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.6 | GO:0012505 | endomembrane system(GO:0012505) |
| 0.0 | 0.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.3 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 1.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 3.9 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 1.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.0 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 1.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 3.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 2.1 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 1.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 1.4 | 4.1 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 1.1 | 4.4 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 1.0 | 6.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.9 | 4.3 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.8 | 2.5 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.8 | 2.3 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.7 | 3.7 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.7 | 2.2 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.7 | 2.2 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.7 | 2.2 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.6 | 4.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.6 | 10.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.5 | 3.8 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.5 | 2.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.4 | 1.3 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.4 | 4.7 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.4 | 19.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.3 | 1.0 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.3 | 1.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.3 | 1.7 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.3 | 3.7 | GO:0046790 | virion binding(GO:0046790) |
| 0.3 | 1.0 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) oncostatin-M receptor activity(GO:0004924) interleukin-6 binding(GO:0019981) |
| 0.3 | 0.9 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.3 | 1.5 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.3 | 1.8 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.3 | 0.9 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.3 | 2.7 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.3 | 2.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.3 | 12.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 1.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.3 | 0.9 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.3 | 0.9 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.3 | 2.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.3 | 2.3 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.3 | 1.9 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.3 | 1.0 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.3 | 1.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.2 | 1.5 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.2 | 1.7 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.2 | 1.8 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.2 | 0.7 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.2 | 8.9 | GO:0016896 | exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.2 | 1.2 | GO:0050542 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.2 | 4.7 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.2 | 1.2 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.2 | 2.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.2 | 2.9 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 6.9 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.2 | 0.6 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.2 | 5.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 1.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.2 | 2.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 2.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 3.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 1.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 0.9 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.2 | 1.0 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.2 | 1.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 1.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 1.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.6 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.4 | GO:0004769 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 1.6 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 6.6 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 0.6 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 2.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.5 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 0.7 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 1.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 2.6 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 0.4 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 1.9 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 2.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.5 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 1.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.1 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 1.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.7 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.8 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 0.6 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 1.9 | GO:0032794 | nitric-oxide synthase regulator activity(GO:0030235) GTPase activating protein binding(GO:0032794) |
| 0.1 | 1.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.9 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 4.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 4.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 3.0 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.3 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 0.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 3.0 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 0.9 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 1.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 1.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 4.4 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 2.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 2.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 1.6 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 0.4 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 2.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.6 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 0.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 3.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.7 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.2 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.4 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 2.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.5 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 1.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 1.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.0 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.6 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.6 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 3.9 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 1.6 | GO:0005126 | cytokine receptor binding(GO:0005126) |
| 0.0 | 0.5 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.8 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 1.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 12.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 8.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 2.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.8 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 3.4 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 4.8 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.2 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.4 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.1 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.0 | 1.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.0 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.4 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.6 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 0.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 12.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.3 | 3.9 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 7.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 5.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.2 | 5.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 7.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 12.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 5.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 7.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 3.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 5.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 4.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 4.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 2.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 1.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 11.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 0.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.9 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.0 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 2.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 5.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.9 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.6 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 1.6 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 2.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.6 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 16.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.4 | 8.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 12.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 3.7 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.3 | 2.8 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.3 | 18.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.3 | 2.9 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.3 | 4.9 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.2 | 1.9 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 1.7 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.2 | 1.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 3.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 2.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 2.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 6.2 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 2.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 3.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 9.4 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.1 | 2.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.4 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 3.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 4.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 2.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 0.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 3.3 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.1 | 2.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 2.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 0.9 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 3.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 1.7 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 2.4 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 3.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 2.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 4.3 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.1 | 1.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 3.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.2 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.7 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 4.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 2.4 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.2 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 2.0 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.1 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 2.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 6.7 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 1.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 1.8 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.0 | 0.6 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.6 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |