Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
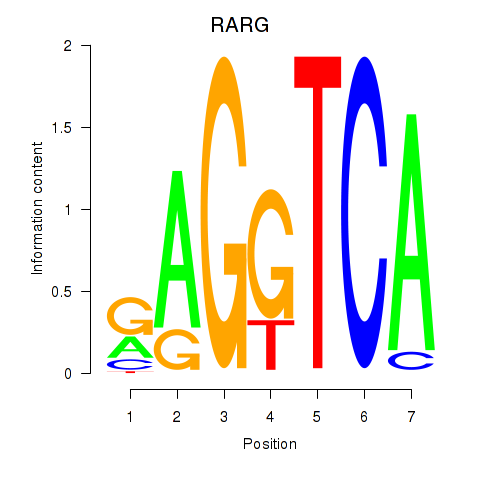
Results for RARG
Z-value: 1.63
Transcription factors associated with RARG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RARG
|
ENSG00000172819.12 | retinoic acid receptor gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RARG | hg19_v2_chr12_-_53614155_53614197, hg19_v2_chr12_-_53614043_53614154 | 0.05 | 5.0e-01 | Click! |
Activity profile of RARG motif
Sorted Z-values of RARG motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RARG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.1 | 39.4 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 9.1 | 64.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 7.0 | 35.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 6.6 | 19.7 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 6.4 | 25.5 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 5.0 | 15.0 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 4.7 | 28.4 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 4.6 | 13.7 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 4.5 | 13.6 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 4.5 | 81.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 4.5 | 49.3 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 4.4 | 13.2 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 4.4 | 17.4 | GO:0009956 | radial pattern formation(GO:0009956) |
| 4.2 | 25.0 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 4.1 | 20.6 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 4.1 | 16.4 | GO:0046502 | uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 3.9 | 15.5 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 3.8 | 11.5 | GO:1904647 | response to rotenone(GO:1904647) |
| 3.8 | 22.9 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 3.8 | 11.4 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 3.7 | 14.8 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 3.5 | 3.5 | GO:0048026 | positive regulation of RNA splicing(GO:0033120) positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 3.2 | 19.1 | GO:0030421 | defecation(GO:0030421) |
| 3.2 | 25.2 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 3.0 | 18.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 3.0 | 12.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 2.9 | 8.8 | GO:1900158 | negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 2.9 | 45.6 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 2.8 | 11.4 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 2.8 | 11.1 | GO:0046108 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 2.6 | 13.2 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 2.6 | 10.5 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 2.6 | 18.3 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 2.6 | 13.0 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 2.6 | 2.6 | GO:0001935 | endothelial cell proliferation(GO:0001935) |
| 2.5 | 7.5 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 2.5 | 9.9 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 2.4 | 9.8 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 2.4 | 14.5 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 2.4 | 9.7 | GO:0043335 | protein unfolding(GO:0043335) |
| 2.3 | 27.6 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 2.2 | 11.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 2.2 | 17.8 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 2.2 | 6.6 | GO:2000276 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 2.1 | 12.8 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 2.1 | 12.8 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 2.1 | 17.0 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 2.1 | 20.8 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 2.1 | 8.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 2.0 | 2.0 | GO:0043317 | regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 2.0 | 12.1 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 2.0 | 5.9 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 2.0 | 5.9 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 1.9 | 40.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 1.9 | 11.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 1.9 | 3.7 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 1.8 | 27.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 1.8 | 5.5 | GO:1902623 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 1.8 | 21.8 | GO:0006108 | malate metabolic process(GO:0006108) |
| 1.8 | 5.4 | GO:0060086 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) circadian temperature homeostasis(GO:0060086) |
| 1.8 | 5.4 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 1.7 | 8.7 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 1.7 | 8.7 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 1.7 | 8.7 | GO:0035900 | response to isolation stress(GO:0035900) |
| 1.7 | 3.5 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 1.7 | 6.9 | GO:0019046 | release from viral latency(GO:0019046) |
| 1.7 | 13.8 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 1.6 | 6.5 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 1.6 | 1.6 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 1.6 | 11.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 1.6 | 1.6 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 1.6 | 12.4 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 1.5 | 4.6 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 1.5 | 4.6 | GO:0060166 | olfactory pit development(GO:0060166) |
| 1.5 | 9.2 | GO:0009624 | response to nematode(GO:0009624) |
| 1.5 | 26.5 | GO:0050812 | regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 1.5 | 13.2 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 1.4 | 4.3 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 1.4 | 11.5 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 1.4 | 2.9 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 1.4 | 8.5 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 1.4 | 5.5 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 1.4 | 4.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 1.4 | 5.5 | GO:0045964 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 1.4 | 6.8 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 1.4 | 6.8 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 1.3 | 4.0 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 1.3 | 4.0 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 1.3 | 18.6 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 1.3 | 4.0 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 1.3 | 5.2 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 1.3 | 9.2 | GO:0015677 | copper ion import(GO:0015677) |
| 1.3 | 2.6 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 1.3 | 7.7 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 1.3 | 29.4 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 1.2 | 3.7 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 1.2 | 2.4 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 1.1 | 14.9 | GO:0016559 | peroxisome fission(GO:0016559) |
| 1.1 | 8.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 1.1 | 4.5 | GO:0030242 | pexophagy(GO:0030242) |
| 1.1 | 18.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 1.1 | 11.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 1.1 | 8.8 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.1 | 5.4 | GO:0061552 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) blood vessel endothelial cell differentiation(GO:0060837) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) ganglion morphogenesis(GO:0061552) blood vessel endothelial cell fate specification(GO:0097101) endothelial tip cell fate specification(GO:0097102) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 1.1 | 7.6 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 1.1 | 2.1 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 1.1 | 12.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 1.1 | 5.3 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 1.1 | 63.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 1.0 | 4.2 | GO:0021539 | subthalamus development(GO:0021539) |
| 1.0 | 4.1 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 1.0 | 20.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 1.0 | 3.1 | GO:0032242 | regulation of nucleoside transport(GO:0032242) negative regulation of neurotrophin production(GO:0032900) |
| 1.0 | 4.1 | GO:0002884 | type IV hypersensitivity(GO:0001806) regulation of type IV hypersensitivity(GO:0001807) negative regulation of hypersensitivity(GO:0002884) |
| 1.0 | 6.0 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 1.0 | 3.0 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 1.0 | 15.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 1.0 | 8.6 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 1.0 | 3.8 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.9 | 4.7 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.9 | 8.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.9 | 16.8 | GO:0006743 | ubiquinone metabolic process(GO:0006743) |
| 0.9 | 2.8 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.9 | 5.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.9 | 8.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.9 | 6.4 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.9 | 3.6 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.9 | 4.4 | GO:1901909 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.9 | 16.6 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.9 | 11.4 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.9 | 7.0 | GO:0014733 | regulation of skeletal muscle adaptation(GO:0014733) |
| 0.9 | 8.7 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.9 | 2.6 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.9 | 4.3 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.9 | 2.6 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.9 | 1.7 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.8 | 3.4 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.8 | 3.4 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.8 | 5.9 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.8 | 11.5 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.8 | 3.3 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.8 | 4.1 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.8 | 2.4 | GO:0015729 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.8 | 9.7 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.8 | 2.4 | GO:0002752 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.8 | 3.2 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.8 | 3.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.8 | 34.7 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.8 | 0.8 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.8 | 1.5 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.8 | 7.0 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.8 | 6.9 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.7 | 9.7 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.7 | 3.7 | GO:0035989 | tendon development(GO:0035989) |
| 0.7 | 5.2 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.7 | 3.7 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.7 | 3.0 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.7 | 2.2 | GO:0090521 | regulation of embryonic cell shape(GO:0016476) glomerular visceral epithelial cell migration(GO:0090521) |
| 0.7 | 1.5 | GO:0044571 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) [2Fe-2S] cluster assembly(GO:0044571) |
| 0.7 | 2.9 | GO:1902031 | regulation of pentose-phosphate shunt(GO:0043456) regulation of NADP metabolic process(GO:1902031) |
| 0.7 | 3.6 | GO:0032907 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) negative regulation of lung blood pressure(GO:0061767) |
| 0.7 | 19.1 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.7 | 2.8 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.7 | 12.4 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.7 | 2.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.7 | 2.8 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.7 | 4.7 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.7 | 2.0 | GO:0060595 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) mammary gland specification(GO:0060594) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.7 | 1.3 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.7 | 8.1 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.7 | 2.7 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.7 | 2.0 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.7 | 29.7 | GO:0061718 | NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.7 | 9.9 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.7 | 3.9 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.6 | 3.2 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.6 | 3.1 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.6 | 2.5 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.6 | 1.8 | GO:1902256 | endocardial cushion fusion(GO:0003274) apoptotic process involved in outflow tract morphogenesis(GO:0003275) negative regulation of alkaline phosphatase activity(GO:0010693) positive regulation of catagen(GO:0051795) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.6 | 9.1 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.6 | 1.8 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.6 | 1.8 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.6 | 11.7 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.6 | 13.2 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.6 | 9.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.6 | 1.7 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.6 | 1.7 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.6 | 1.7 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.5 | 1.6 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.5 | 6.0 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.5 | 4.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.5 | 3.7 | GO:1903977 | positive regulation of glial cell migration(GO:1903977) |
| 0.5 | 4.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.5 | 2.1 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.5 | 1.6 | GO:0071609 | chemokine (C-C motif) ligand 5 production(GO:0071609) |
| 0.5 | 2.6 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.5 | 4.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.5 | 1.5 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.5 | 2.5 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.5 | 2.5 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.5 | 4.5 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.5 | 1.5 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.5 | 2.0 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.5 | 2.9 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.5 | 15.5 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.5 | 11.1 | GO:0042749 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.5 | 0.5 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.5 | 6.7 | GO:0090343 | positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 0.5 | 1.9 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.5 | 9.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.5 | 1.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.5 | 3.7 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.5 | 7.8 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.5 | 2.3 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.5 | 4.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.5 | 1.8 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.5 | 11.7 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.5 | 4.5 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.4 | 0.9 | GO:0046440 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.4 | 0.4 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.4 | 1.8 | GO:0090296 | regulation of mitochondrial DNA replication(GO:0090296) |
| 0.4 | 1.7 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.4 | 3.0 | GO:1903286 | regulation of potassium ion import(GO:1903286) |
| 0.4 | 3.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.4 | 0.9 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 0.4 | 1.7 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.4 | 0.4 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.4 | 2.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.4 | 1.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.4 | 2.1 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.4 | 7.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.4 | 4.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.4 | 2.5 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.4 | 1.7 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.4 | 16.0 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.4 | 6.5 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.4 | 2.4 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.4 | 6.1 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.4 | 2.0 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.4 | 2.4 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.4 | 2.4 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.4 | 6.0 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.4 | 6.7 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.4 | 1.6 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.4 | 1.6 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.4 | 11.9 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.4 | 5.7 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.4 | 1.5 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.4 | 1.9 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.4 | 3.8 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.4 | 20.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.4 | 3.0 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.4 | 2.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.4 | 9.3 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.4 | 7.0 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.4 | 8.4 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.4 | 10.2 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.4 | 0.7 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.4 | 3.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.4 | 2.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.4 | 1.8 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.4 | 1.1 | GO:0051037 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.4 | 1.8 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.3 | 2.4 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.3 | 6.2 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.3 | 1.4 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.3 | 2.4 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.3 | 4.1 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.3 | 4.0 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.3 | 3.0 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.3 | 5.6 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.3 | 0.7 | GO:0090175 | establishment of planar polarity(GO:0001736) establishment of tissue polarity(GO:0007164) Wnt signaling pathway, planar cell polarity pathway(GO:0060071) regulation of establishment of planar polarity(GO:0090175) |
| 0.3 | 7.6 | GO:0050718 | positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.3 | 3.6 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.3 | 7.8 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.3 | 2.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.3 | 1.3 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.3 | 11.9 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.3 | 7.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.3 | 4.8 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.3 | 1.6 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.3 | 3.5 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.3 | 6.6 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.3 | 4.1 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.3 | 1.9 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.3 | 3.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.3 | 9.7 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.3 | 6.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.3 | 0.9 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.3 | 8.3 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.3 | 8.8 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.3 | 1.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.3 | 3.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.3 | 1.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.3 | 12.9 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.3 | 1.7 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.3 | 11.6 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.3 | 8.9 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.3 | 3.7 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.3 | 2.0 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.3 | 1.7 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.3 | 1.1 | GO:0001880 | Mullerian duct regression(GO:0001880) regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.3 | 0.8 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.3 | 1.9 | GO:0035898 | parathyroid hormone secretion(GO:0035898) post-embryonic body morphogenesis(GO:0040032) regulation of parathyroid hormone secretion(GO:2000828) |
| 0.3 | 2.5 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.3 | 3.0 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.3 | 1.4 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.3 | 0.8 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) intestinal epithelial cell maturation(GO:0060574) |
| 0.3 | 9.8 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.3 | 0.8 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 5.0 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.3 | 17.3 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.3 | 5.4 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.3 | 4.3 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.3 | 2.0 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.3 | 2.0 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.2 | 3.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 2.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.2 | 5.2 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.2 | 1.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 2.7 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.2 | 0.5 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.2 | 2.7 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.2 | 2.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 | 6.0 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.2 | 3.1 | GO:0035878 | nail development(GO:0035878) |
| 0.2 | 6.9 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.2 | 2.1 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.2 | 1.9 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.2 | 4.5 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.2 | 3.3 | GO:2000404 | regulation of T cell migration(GO:2000404) |
| 0.2 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.2 | 1.2 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.2 | 3.7 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.2 | 10.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 8.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.2 | 0.7 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.2 | 2.8 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 3.9 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.2 | 1.8 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.2 | 2.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.2 | 0.5 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.2 | 2.0 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.2 | 17.8 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.2 | 0.7 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.2 | 1.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 15.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.2 | 10.3 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.2 | 6.1 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.2 | 1.3 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.2 | 3.0 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.2 | 2.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.2 | 3.4 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 1.0 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 1.0 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.2 | 1.8 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.2 | 0.8 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.2 | 2.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 0.8 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.2 | 2.8 | GO:0060996 | dendritic spine development(GO:0060996) |
| 0.2 | 2.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 2.5 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.2 | 2.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.2 | 2.1 | GO:0072678 | T cell migration(GO:0072678) |
| 0.2 | 2.6 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.2 | 0.6 | GO:0086098 | angiotensin-activated signaling pathway involved in heart process(GO:0086098) |
| 0.2 | 0.7 | GO:0042335 | cuticle development(GO:0042335) |
| 0.2 | 1.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 0.5 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.2 | 1.9 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.2 | 8.0 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.2 | 3.4 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.2 | 6.4 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.2 | 3.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 2.8 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.2 | 3.0 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.2 | 0.7 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.2 | 0.7 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.2 | 0.7 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.2 | 1.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.2 | 1.0 | GO:0033014 | tetrapyrrole biosynthetic process(GO:0033014) |
| 0.2 | 2.1 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.2 | 0.6 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.2 | 1.6 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.2 | 2.5 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.2 | 1.1 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.2 | 2.3 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.2 | 0.5 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.2 | 1.5 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.2 | 5.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 4.8 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.1 | 0.9 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 1.0 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.9 | GO:1990000 | amyloid fibril formation(GO:1990000) |
| 0.1 | 1.9 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.1 | 13.0 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.1 | 1.6 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 0.9 | GO:0071907 | determination of pancreatic left/right asymmetry(GO:0035469) determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.1 | 1.7 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 3.9 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 0.9 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 | 5.3 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 5.3 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.1 | 1.3 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 1.2 | GO:0034391 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.1 | 0.7 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 1.8 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.1 | 0.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 1.8 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.8 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 6.7 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.1 | 1.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 3.8 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 3.4 | GO:1902400 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.1 | 4.7 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.1 | 1.2 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.1 | 1.9 | GO:0061213 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 8.7 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.1 | 6.7 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.1 | 2.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 1.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.9 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 4.6 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 4.4 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 1.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.6 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.5 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 2.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 14.5 | GO:0007163 | establishment or maintenance of cell polarity(GO:0007163) |
| 0.1 | 4.0 | GO:0048048 | embryonic eye morphogenesis(GO:0048048) |
| 0.1 | 3.8 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 2.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 4.4 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.1 | 0.5 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 1.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.9 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.1 | 3.0 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 13.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 3.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 2.5 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 0.4 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 4.5 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 1.9 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 3.2 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 1.7 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 1.6 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.7 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 0.3 | GO:0097278 | complement-dependent cytotoxicity(GO:0097278) regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.1 | 1.6 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.4 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.1 | 1.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.9 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 | 0.2 | GO:0032470 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 2.2 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.1 | 0.4 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.1 | 1.4 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.1 | 0.6 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 1.1 | GO:0051931 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.1 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 1.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.3 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 | 1.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 1.7 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 2.2 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 3.3 | GO:0090114 | COPII-coated vesicle budding(GO:0090114) |
| 0.1 | 0.8 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 3.0 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 1.1 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.1 | 3.7 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 0.4 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 1.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.6 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.1 | 0.1 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 4.8 | GO:0017156 | calcium ion regulated exocytosis(GO:0017156) |
| 0.1 | 2.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.5 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.4 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.6 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.0 | 0.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 1.2 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.6 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 2.2 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 2.3 | GO:1903318 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 | 0.6 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.6 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.4 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 1.8 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.3 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.2 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.3 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.0 | 1.5 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.5 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.2 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 6.3 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 1.1 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 11.5 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 1.1 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 1.2 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 1.3 | GO:0009791 | post-embryonic development(GO:0009791) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 1.7 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.8 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.7 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.1 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.1 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.0 | 0.6 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 1.1 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.5 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.3 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.0 | GO:2000698 | positive regulation of epithelial cell differentiation involved in kidney development(GO:2000698) |
| 0.0 | 0.7 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.0 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.1 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.3 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.3 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.9 | GO:0007017 | microtubule-based process(GO:0007017) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.5 | 25.5 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 5.7 | 5.7 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 5.0 | 79.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 4.8 | 95.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 4.7 | 28.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 3.5 | 10.6 | GO:0072534 | perineuronal net(GO:0072534) |
| 3.1 | 15.6 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 2.7 | 16.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 2.5 | 35.1 | GO:0097227 | sperm annulus(GO:0097227) |
| 2.5 | 7.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 2.5 | 9.8 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 2.2 | 11.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 2.2 | 15.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 2.1 | 10.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 2.1 | 16.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 2.0 | 7.8 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 2.0 | 13.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.9 | 17.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 1.9 | 5.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 1.8 | 17.6 | GO:0045180 | basal cortex(GO:0045180) |
| 1.7 | 16.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 1.6 | 49.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 1.6 | 22.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 1.6 | 6.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 1.6 | 10.9 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 1.5 | 4.5 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 1.5 | 3.0 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 1.4 | 2.9 | GO:0012505 | endomembrane system(GO:0012505) |
| 1.3 | 10.8 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 1.3 | 4.0 | GO:0043291 | RAVE complex(GO:0043291) |
| 1.3 | 7.6 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 1.3 | 16.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 1.2 | 13.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 1.2 | 8.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 1.2 | 6.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 1.2 | 15.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 1.2 | 6.9 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 1.2 | 24.2 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 1.1 | 11.5 | GO:0097427 | microtubule bundle(GO:0097427) |
| 1.1 | 10.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 1.1 | 9.9 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 1.1 | 12.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.1 | 6.5 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 1.1 | 10.5 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 1.0 | 2.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 1.0 | 26.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 1.0 | 5.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 1.0 | 3.8 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 1.0 | 14.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.9 | 13.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.9 | 3.7 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.9 | 55.1 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.9 | 2.8 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.9 | 9.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.9 | 16.8 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.8 | 1.7 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.8 | 6.6 | GO:0060203 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.8 | 4.0 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.8 | 18.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.8 | 3.9 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.8 | 35.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.8 | 56.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.8 | 11.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.7 | 16.2 | GO:0042641 | actomyosin(GO:0042641) |
| 0.7 | 12.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.6 | 3.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.6 | 8.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.6 | 4.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.6 | 6.8 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.6 | 7.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.6 | 16.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.6 | 3.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.6 | 1.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.6 | 23.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.6 | 3.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.5 | 10.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.5 | 24.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.5 | 12.1 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.5 | 18.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.5 | 10.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.5 | 2.0 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.5 | 3.9 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.5 | 10.5 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.5 | 15.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.5 | 26.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.5 | 5.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.5 | 5.6 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.5 | 4.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.5 | 95.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.5 | 3.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.5 | 4.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.5 | 9.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.4 | 3.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.4 | 9.7 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.4 | 1.7 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.4 | 3.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.4 | 1.6 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.4 | 8.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.4 | 2.0 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.4 | 2.4 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.4 | 3.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.4 | 1.6 | GO:0031417 | NatC complex(GO:0031417) |
| 0.4 | 15.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.4 | 3.9 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.4 | 11.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.4 | 1.5 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.4 | 1.9 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.4 | 1.8 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.4 | 1.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.4 | 5.4 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.4 | 4.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.3 | 2.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.3 | 3.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 10.7 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.3 | 17.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.3 | 3.6 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.3 | 4.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 3.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 1.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.3 | 3.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.3 | 4.7 | GO:0010369 | chromocenter(GO:0010369) |
| 0.3 | 7.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.3 | 3.0 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.3 | 14.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.3 | 10.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 4.6 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.3 | 23.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.3 | 1.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.3 | 5.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 3.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 2.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 1.0 | GO:0045179 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) apical cortex(GO:0045179) |
| 0.2 | 5.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 4.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 5.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.2 | 4.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 1.6 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 13.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 54.3 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.2 | 3.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 12.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 4.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 2.7 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.2 | 2.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.2 | 1.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 0.7 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 0.9 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.2 | 3.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 1.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 20.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 23.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 1.6 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.2 | 4.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 0.5 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.2 | 3.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.2 | 3.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 3.0 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.2 | 9.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 3.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 4.6 | GO:0031256 | leading edge membrane(GO:0031256) |
| 0.1 | 2.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.6 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 2.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 6.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 1.7 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.8 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 10.1 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 1.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 4.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 4.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 7.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.0 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.7 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 6.0 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 3.0 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 3.1 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 0.8 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 2.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 26.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 1.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 1.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 1.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 3.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 2.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.3 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 0.5 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.1 | 2.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 2.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 3.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 2.9 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 1.6 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 2.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 1.0 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 3.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 8.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 49.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.1 | 2.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.4 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 6.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.4 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 3.0 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 3.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 2.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 2.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 9.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 3.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.6 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 1.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 7.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 2.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.4 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 0.2 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.3 | GO:0036126 | sperm flagellum(GO:0036126) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.3 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 5.4 | 16.1 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 5.2 | 83.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 5.2 | 15.6 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 4.7 | 28.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 4.4 | 13.2 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 3.5 | 10.5 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 2.9 | 14.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 2.9 | 14.6 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 2.9 | 11.6 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 2.8 | 11.1 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 2.6 | 7.8 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 2.6 | 5.2 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 2.5 | 7.5 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 2.5 | 77.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 2.4 | 14.6 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 2.4 | 26.0 | GO:0015266 | protein channel activity(GO:0015266) |
| 2.2 | 6.5 | GO:0048030 | disaccharide binding(GO:0048030) |
| 2.1 | 42.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 2.1 | 12.8 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 2.1 | 12.7 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 2.1 | 10.5 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 2.1 | 20.8 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 2.0 | 9.8 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 1.9 | 19.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.9 | 21.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 1.9 | 5.7 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 1.9 | 14.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.8 | 14.5 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 1.8 | 25.0 | GO:0031432 | titin binding(GO:0031432) |
| 1.7 | 8.7 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 1.7 | 12.0 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 1.7 | 20.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 1.6 | 4.9 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 1.6 | 11.2 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 1.6 | 19.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 1.6 | 45.2 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 1.5 | 10.3 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 1.5 | 8.8 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.4 | 2.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 1.4 | 26.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 1.4 | 4.1 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 1.4 | 13.7 | GO:0030911 | TPR domain binding(GO:0030911) |
| 1.3 | 5.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 1.3 | 11.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 1.3 | 7.6 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 1.3 | 8.8 | GO:0016015 | morphogen activity(GO:0016015) |
| 1.2 | 3.7 | GO:0070052 | collagen V binding(GO:0070052) |
| 1.2 | 3.7 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 1.2 | 7.2 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 1.2 | 3.6 | GO:0004040 | amidase activity(GO:0004040) |
| 1.2 | 10.6 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 1.2 | 10.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 1.2 | 4.6 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 1.1 | 10.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 1.1 | 8.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 1.1 | 4.4 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 1.1 | 7.6 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 1.1 | 10.6 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 1.1 | 5.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 1.0 | 6.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.0 | 3.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 1.0 | 4.1 | GO:0052812 | phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 1.0 | 6.0 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 1.0 | 3.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 1.0 | 5.0 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 1.0 | 5.8 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 1.0 | 6.8 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.9 | 13.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.9 | 8.5 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.9 | 6.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.9 | 3.7 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.9 | 11.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.9 | 10.9 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.9 | 4.5 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.9 | 5.4 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.9 | 4.5 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.9 | 2.7 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.9 | 4.4 | GO:0052846 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.9 | 9.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.9 | 10.6 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.9 | 4.3 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.8 | 4.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.8 | 8.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.8 | 3.3 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.8 | 20.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.8 | 17.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.8 | 21.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.8 | 2.4 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.8 | 8.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.8 | 3.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.8 | 35.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.8 | 5.4 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.7 | 12.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.7 | 3.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.7 | 2.9 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.7 | 13.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.7 | 2.1 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.7 | 3.6 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.7 | 2.8 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.7 | 4.2 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.7 | 11.0 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.7 | 2.7 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.7 | 6.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.7 | 2.6 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.7 | 2.6 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.7 | 2.0 | GO:0019961 | interferon binding(GO:0019961) |
| 0.7 | 3.9 | GO:0015288 | porin activity(GO:0015288) |
| 0.6 | 0.6 | GO:0032557 | pyrimidine ribonucleotide binding(GO:0032557) |
| 0.6 | 17.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.6 | 1.9 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.6 | 8.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.6 | 6.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.6 | 6.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.6 | 3.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.6 | 6.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.6 | 1.8 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.6 | 2.4 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.6 | 4.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.6 | 10.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.6 | 3.5 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.6 | 1.7 | GO:0031692 | alpha-1A adrenergic receptor binding(GO:0031691) alpha-1B adrenergic receptor binding(GO:0031692) follicle-stimulating hormone receptor binding(GO:0031762) V2 vasopressin receptor binding(GO:0031896) |
| 0.6 | 3.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.6 | 6.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.6 | 15.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.6 | 7.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.6 | 3.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.5 | 1.6 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.5 | 1.6 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.5 | 21.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.5 | 1.6 | GO:0004515 | nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.5 | 2.7 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.5 | 2.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.5 | 2.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.5 | 1.6 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.5 | 7.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.5 | 3.1 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.5 | 3.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.5 | 3.1 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.5 | 1.5 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.5 | 17.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.5 | 4.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.5 | 3.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.5 | 46.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.5 | 12.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.5 | 7.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.5 | 10.7 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.5 | 10.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.5 | 18.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.5 | 1.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.5 | 7.7 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.4 | 1.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.4 | 1.3 | GO:0016730 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.4 | 10.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.4 | 1.7 | GO:0047298 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.4 | 3.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.4 | 44.3 | GO:0003774 | motor activity(GO:0003774) |
| 0.4 | 2.5 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.4 | 6.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.4 | 1.6 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.4 | 4.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 2.4 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.4 | 1.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.4 | 1.6 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.4 | 7.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.4 | 5.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.4 | 16.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.4 | 1.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.4 | 3.9 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.4 | 0.8 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.4 | 3.0 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.4 | 1.5 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.4 | 3.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.4 | 10.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.4 | 49.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.4 | 2.9 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.4 | 6.0 | GO:0031005 | filamin binding(GO:0031005) |
| 0.4 | 1.4 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.3 | 0.7 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.3 | 9.0 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.3 | 14.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.3 | 15.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.3 | 1.7 | GO:0016301 | kinase activity(GO:0016301) |
| 0.3 | 2.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.3 | 5.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.3 | 6.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.3 | 1.3 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.3 | 11.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.3 | 7.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.3 | 15.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.3 | 16.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.3 | 13.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.3 | 1.6 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.3 | 0.9 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.3 | 7.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.3 | 1.3 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.3 | 18.1 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.3 | 4.3 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.3 | 7.0 | GO:0046961 | hydrogen-exporting ATPase activity(GO:0036442) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.3 | 1.8 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.3 | 0.9 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.3 | 4.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.3 | 1.8 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.3 | 1.8 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.3 | 1.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.3 | 11.4 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.3 | 1.7 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.3 | 12.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.3 | 8.6 | GO:0001848 | complement binding(GO:0001848) |
| 0.3 | 1.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.3 | 1.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.3 | 23.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.3 | 1.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.3 | 7.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 3.5 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.3 | 2.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.3 | 2.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.3 | 0.8 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.3 | 4.0 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.2 | 3.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.2 | 2.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 3.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 0.7 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.2 | 3.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 1.7 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 1.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 2.5 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.2 | 4.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 2.7 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.2 | 1.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 1.8 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 1.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.2 | 0.7 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.2 | 5.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 11.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 2.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 1.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.2 | 0.8 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.2 | 9.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 4.8 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.2 | 3.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 1.0 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 1.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 1.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 4.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 2.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 2.0 | GO:0009374 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.2 | 2.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 1.2 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.2 | 1.6 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 2.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 2.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.2 | 1.4 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.2 | 1.8 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 7.6 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.2 | 36.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.2 | 5.0 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 15.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 5.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 1.9 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.2 | 3.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.2 | 3.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 1.9 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 8.7 | GO:0033293 | monocarboxylic acid binding(GO:0033293) |
| 0.2 | 3.7 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.2 | 2.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 0.7 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.2 | 1.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 1.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 0.5 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.2 | 1.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 1.4 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.2 | 0.5 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.2 | 0.6 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 2.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 4.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.2 | 3.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.2 | 2.4 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 5.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.9 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 2.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 1.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 9.9 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 2.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 2.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 0.5 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.2 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 5.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 3.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.8 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.1 | 6.2 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.1 | 0.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.9 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 16.8 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 1.9 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 3.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.6 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 1.0 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 3.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 4.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 2.0 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 1.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 14.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 0.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 5.0 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.1 | 1.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 3.9 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 5.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 1.5 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 1.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.9 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 0.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.9 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 1.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 2.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 23.9 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 3.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.6 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.4 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 1.0 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.1 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.7 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 1.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.9 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.2 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.1 | 2.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.3 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 0.4 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 2.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 2.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 1.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.7 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 1.6 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.7 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 1.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 1.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 2.7 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 3.8 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 2.3 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 2.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 3.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 7.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.4 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.7 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 2.2 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 1.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 2.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.6 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 2.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 2.1 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.1 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.7 | GO:0022832 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.0 | 0.2 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 1.0 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.4 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.1 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 0.1 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.8 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.0 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 31.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 1.1 | 82.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.8 | 10.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.8 | 19.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.7 | 46.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.7 | 7.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.6 | 14.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.6 | 50.2 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.6 | 17.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.6 | 34.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.5 | 21.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.5 | 11.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.5 | 6.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.5 | 21.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.5 | 9.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.4 | 5.8 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.4 | 13.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.4 | 2.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.4 | 15.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.4 | 10.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 10.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.3 | 15.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.3 | 10.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.3 | 25.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.3 | 16.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.3 | 25.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.3 | 10.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.3 | 4.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 8.9 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.2 | 6.3 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.2 | 8.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 15.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 4.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 3.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 4.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.2 | 2.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 1.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 1.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.2 | 0.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.2 | 2.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 6.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 2.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 3.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 4.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 3.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 11.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 44.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 3.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 2.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 20.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 0.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 5.3 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 1.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 4.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 4.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 18.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 0.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 4.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 0.9 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 2.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 4.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 4.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.0 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 3.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 51.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 1.6 | 26.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 1.5 | 83.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 1.5 | 16.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 1.4 | 120.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 1.2 | 31.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 1.1 | 19.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 1.0 | 12.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 1.0 | 92.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.9 | 16.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.9 | 6.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.9 | 14.9 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.9 | 20.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.8 | 5.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.8 | 26.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.8 | 49.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.8 | 17.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.7 | 9.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.7 | 12.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.6 | 41.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.6 | 13.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.6 | 8.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.6 | 21.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.6 | 9.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.6 | 27.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.5 | 12.0 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.5 | 5.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.5 | 10.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.5 | 9.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.5 | 17.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.5 | 8.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.5 | 10.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.5 | 8.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.5 | 11.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.5 | 5.8 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.5 | 10.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.5 | 16.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.5 | 16.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.5 | 15.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.4 | 7.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.4 | 7.9 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.4 | 12.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.4 | 3.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.4 | 10.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.4 | 2.6 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.4 | 9.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.4 | 2.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.3 | 10.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.3 | 9.9 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.3 | 6.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.3 | 6.8 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.3 | 5.0 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.3 | 5.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.3 | 6.7 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.3 | 6.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 4.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.3 | 8.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 5.0 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.3 | 4.6 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.3 | 12.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.3 | 7.4 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.3 | 16.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 5.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 23.8 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.2 | 3.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 2.9 | REACTOME ORC1 REMOVAL FROM CHROMATIN | Genes involved in Orc1 removal from chromatin |
| 0.2 | 16.9 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.2 | 16.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.2 | 4.9 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 9.1 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.2 | 5.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 3.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 17.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 1.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.2 | 3.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 1.4 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.2 | 1.9 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 3.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 5.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 0.5 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.2 | 3.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 4.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 2.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.5 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 1.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 3.1 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 2.0 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 1.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 4.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 5.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 2.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.7 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 1.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.7 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 2.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 4.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 2.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 12.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 3.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.3 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.1 | 1.8 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 0.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.1 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.1 | 1.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 0.9 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 1.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 1.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.2 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |