Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
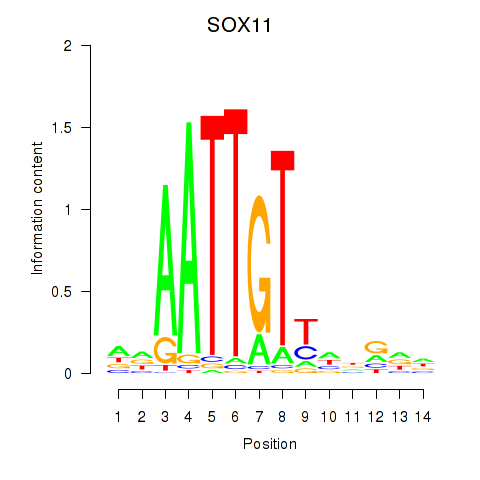
Results for SOX11
Z-value: 0.52
Transcription factors associated with SOX11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX11
|
ENSG00000176887.5 | SRY-box transcription factor 11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX11 | hg19_v2_chr2_+_5832799_5832799 | 0.04 | 5.2e-01 | Click! |
Activity profile of SOX11 motif
Sorted Z-values of SOX11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_64673630 | 7.69 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chrX_-_13835147 | 7.47 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr7_+_98923505 | 7.02 |
ENST00000432884.2
ENST00000262942.5 |
ARPC1A
|
actin related protein 2/3 complex, subunit 1A, 41kDa |
| chr4_-_90759440 | 6.67 |
ENST00000336904.3
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr2_-_176046391 | 5.40 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr2_+_187371440 | 5.22 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr15_+_80733570 | 5.19 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr3_+_69812701 | 4.96 |
ENST00000472437.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr10_-_95242044 | 4.76 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr6_-_116381918 | 4.66 |
ENST00000606080.1
|
FRK
|
fyn-related kinase |
| chr10_-_95241951 | 4.64 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr8_-_13134045 | 4.59 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr6_+_30585486 | 4.52 |
ENST00000259873.4
ENST00000506373.2 |
MRPS18B
|
mitochondrial ribosomal protein S18B |
| chr15_-_37392086 | 4.46 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr2_+_47596287 | 4.17 |
ENST00000263735.4
|
EPCAM
|
epithelial cell adhesion molecule |
| chr17_-_79895097 | 3.92 |
ENST00000402252.2
ENST00000583564.1 ENST00000585244.1 ENST00000337943.5 ENST00000579698.1 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr11_-_95657231 | 3.90 |
ENST00000409459.1
ENST00000352297.7 ENST00000393223.3 ENST00000346299.5 |
MTMR2
|
myotubularin related protein 2 |
| chr1_+_214776516 | 3.83 |
ENST00000366955.3
|
CENPF
|
centromere protein F, 350/400kDa |
| chr15_-_64673665 | 3.72 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr3_-_47950745 | 3.66 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr17_+_6347729 | 3.63 |
ENST00000572447.1
|
FAM64A
|
family with sequence similarity 64, member A |
| chr11_+_12399071 | 3.62 |
ENST00000539723.1
ENST00000550549.1 |
PARVA
|
parvin, alpha |
| chr17_+_6347761 | 3.55 |
ENST00000250056.8
ENST00000571373.1 ENST00000570337.2 ENST00000572595.2 ENST00000576056.1 |
FAM64A
|
family with sequence similarity 64, member A |
| chr17_-_79895154 | 3.46 |
ENST00000405481.4
ENST00000585215.1 ENST00000577624.1 ENST00000403172.4 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr2_-_178128250 | 3.03 |
ENST00000448782.1
ENST00000446151.2 |
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr16_+_67261008 | 2.83 |
ENST00000304800.9
ENST00000563953.1 ENST00000565201.1 |
TMEM208
|
transmembrane protein 208 |
| chr8_-_54752406 | 2.82 |
ENST00000520188.1
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr2_-_225266743 | 2.56 |
ENST00000409685.3
|
FAM124B
|
family with sequence similarity 124B |
| chr2_+_109237717 | 2.55 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr19_+_39989535 | 2.45 |
ENST00000356433.5
|
DLL3
|
delta-like 3 (Drosophila) |
| chr17_-_10017864 | 2.27 |
ENST00000323816.4
|
GAS7
|
growth arrest-specific 7 |
| chr2_-_225266711 | 2.26 |
ENST00000389874.3
|
FAM124B
|
family with sequence similarity 124B |
| chr4_-_170679024 | 2.25 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27 |
| chr12_+_28410128 | 2.21 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr5_+_135496675 | 2.17 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr12_+_50794592 | 2.13 |
ENST00000293618.8
ENST00000429001.3 ENST00000548174.1 ENST00000548697.1 ENST00000548993.1 ENST00000398473.2 ENST00000522085.1 ENST00000518444.1 ENST00000551886.1 |
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr1_-_150669500 | 2.12 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr2_-_37068530 | 2.04 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr5_+_140753444 | 2.00 |
ENST00000517434.1
|
PCDHGA6
|
protocadherin gamma subfamily A, 6 |
| chr18_+_7754957 | 1.99 |
ENST00000400053.4
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr2_-_190044480 | 1.97 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr7_-_82792215 | 1.80 |
ENST00000333891.9
ENST00000423517.2 |
PCLO
|
piccolo presynaptic cytomatrix protein |
| chrX_+_100645812 | 1.78 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr7_-_56118981 | 1.75 |
ENST00000419984.2
ENST00000413218.1 ENST00000424596.1 |
PSPH
|
phosphoserine phosphatase |
| chr8_+_19796381 | 1.72 |
ENST00000524029.1
ENST00000522701.1 ENST00000311322.8 |
LPL
|
lipoprotein lipase |
| chr6_-_11779014 | 1.70 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr20_-_7921090 | 1.68 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr15_+_49447947 | 1.64 |
ENST00000327171.3
ENST00000560654.1 |
GALK2
|
galactokinase 2 |
| chr6_+_26183958 | 1.64 |
ENST00000356530.3
|
HIST1H2BE
|
histone cluster 1, H2be |
| chr10_-_127505167 | 1.62 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr6_-_47277634 | 1.59 |
ENST00000296861.2
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr3_-_133380731 | 1.57 |
ENST00000260810.5
|
TOPBP1
|
topoisomerase (DNA) II binding protein 1 |
| chr8_+_104831554 | 1.57 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr17_-_34344991 | 1.55 |
ENST00000591423.1
|
CCL23
|
chemokine (C-C motif) ligand 23 |
| chr1_-_205744205 | 1.42 |
ENST00000446390.2
|
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr4_+_183164574 | 1.41 |
ENST00000511685.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chrX_-_109590174 | 1.36 |
ENST00000372054.1
|
GNG5P2
|
guanine nucleotide binding protein (G protein), gamma 5 pseudogene 2 |
| chr6_-_11779174 | 1.35 |
ENST00000379413.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr12_-_10955226 | 1.32 |
ENST00000240687.2
|
TAS2R7
|
taste receptor, type 2, member 7 |
| chr3_-_107777208 | 1.28 |
ENST00000398258.3
|
CD47
|
CD47 molecule |
| chr1_-_115292591 | 1.26 |
ENST00000438362.2
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr14_-_36789865 | 1.25 |
ENST00000416007.4
|
MBIP
|
MAP3K12 binding inhibitory protein 1 |
| chr5_+_148521381 | 1.23 |
ENST00000504238.1
|
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr8_+_105352050 | 1.20 |
ENST00000297581.2
|
DCSTAMP
|
dendrocyte expressed seven transmembrane protein |
| chr13_-_30424821 | 1.20 |
ENST00000380680.4
|
UBL3
|
ubiquitin-like 3 |
| chr2_+_138722028 | 1.20 |
ENST00000280096.5
|
HNMT
|
histamine N-methyltransferase |
| chr5_+_169780485 | 1.16 |
ENST00000377360.4
|
KCNIP1
|
Kv channel interacting protein 1 |
| chr8_+_104831472 | 1.14 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr14_-_36789783 | 1.13 |
ENST00000605579.1
ENST00000604336.1 ENST00000359527.7 ENST00000603139.1 ENST00000318473.7 |
MBIP
|
MAP3K12 binding inhibitory protein 1 |
| chr1_-_24469602 | 1.11 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1 |
| chr2_+_138721850 | 1.11 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr17_-_34345002 | 1.03 |
ENST00000293280.2
|
CCL23
|
chemokine (C-C motif) ligand 23 |
| chr1_+_145524891 | 1.01 |
ENST00000369304.3
|
ITGA10
|
integrin, alpha 10 |
| chr4_-_73434498 | 1.00 |
ENST00000286657.4
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr10_+_118350522 | 0.99 |
ENST00000530319.1
ENST00000527980.1 ENST00000471549.1 ENST00000534537.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr3_-_167191814 | 0.88 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr9_+_71986182 | 0.80 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr10_+_5238793 | 0.79 |
ENST00000263126.1
|
AKR1C4
|
aldo-keto reductase family 1, member C4 |
| chr4_-_159080806 | 0.73 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr2_-_20251744 | 0.72 |
ENST00000175091.4
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha |
| chr7_-_122635754 | 0.72 |
ENST00000249284.2
|
TAS2R16
|
taste receptor, type 2, member 16 |
| chr2_+_102928009 | 0.72 |
ENST00000404917.2
ENST00000447231.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr1_+_65730385 | 0.67 |
ENST00000263441.7
ENST00000395325.3 |
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr4_+_72204755 | 0.65 |
ENST00000512686.1
ENST00000340595.3 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr5_+_140248518 | 0.65 |
ENST00000398640.2
|
PCDHA11
|
protocadherin alpha 11 |
| chr11_+_57425209 | 0.64 |
ENST00000533905.1
ENST00000525602.1 ENST00000302731.4 |
CLP1
|
cleavage and polyadenylation factor I subunit 1 |
| chr19_-_14911023 | 0.61 |
ENST00000248073.2
|
OR7C1
|
olfactory receptor, family 7, subfamily C, member 1 |
| chr5_+_173472607 | 0.60 |
ENST00000303177.3
ENST00000519867.1 |
NSG2
|
Neuron-specific protein family member 2 |
| chr8_+_24241789 | 0.59 |
ENST00000256412.4
ENST00000538205.1 |
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr9_-_95166841 | 0.58 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr2_+_102927962 | 0.54 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr21_-_45671014 | 0.53 |
ENST00000436357.1
|
DNMT3L
|
DNA (cytosine-5-)-methyltransferase 3-like |
| chr6_+_29068386 | 0.52 |
ENST00000377171.3
|
OR2J1
|
olfactory receptor, family 2, subfamily J, member 1 (gene/pseudogene) |
| chr6_-_49712123 | 0.52 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr11_+_22688150 | 0.52 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr4_-_110624564 | 0.52 |
ENST00000352981.3
ENST00000265164.2 ENST00000505486.1 |
CASP6
|
caspase 6, apoptosis-related cysteine peptidase |
| chr14_-_20801427 | 0.51 |
ENST00000557665.1
ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr6_-_49604545 | 0.48 |
ENST00000371175.4
ENST00000229810.7 |
RHAG
|
Rh-associated glycoprotein |
| chr4_+_71337834 | 0.45 |
ENST00000304887.5
|
MUC7
|
mucin 7, secreted |
| chr1_+_196621002 | 0.44 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr4_+_169013666 | 0.44 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr3_-_55001115 | 0.43 |
ENST00000493075.1
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr18_+_22040620 | 0.43 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chrX_+_102192200 | 0.42 |
ENST00000218249.5
|
RAB40AL
|
RAB40A, member RAS oncogene family-like |
| chr18_+_22040593 | 0.40 |
ENST00000256906.4
|
HRH4
|
histamine receptor H4 |
| chr3_+_101504200 | 0.39 |
ENST00000422132.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr20_+_35807512 | 0.39 |
ENST00000373622.5
|
RPN2
|
ribophorin II |
| chr11_+_120973375 | 0.37 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr2_-_219157250 | 0.36 |
ENST00000434015.2
ENST00000444183.1 ENST00000420341.1 ENST00000453281.1 ENST00000258412.3 ENST00000440422.1 |
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr12_-_81763127 | 0.36 |
ENST00000541017.1
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr20_-_32308028 | 0.36 |
ENST00000409299.3
ENST00000217398.3 ENST00000344022.3 |
PXMP4
|
peroxisomal membrane protein 4, 24kDa |
| chr8_-_25281747 | 0.35 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr13_-_99630233 | 0.30 |
ENST00000376460.1
ENST00000442173.1 |
DOCK9
|
dedicator of cytokinesis 9 |
| chr17_-_10276319 | 0.29 |
ENST00000252172.4
ENST00000418404.3 |
MYH13
|
myosin, heavy chain 13, skeletal muscle |
| chr3_-_87325728 | 0.28 |
ENST00000350375.2
|
POU1F1
|
POU class 1 homeobox 1 |
| chr10_-_5446786 | 0.28 |
ENST00000479328.1
ENST00000380419.3 |
TUBAL3
|
tubulin, alpha-like 3 |
| chr4_+_110749143 | 0.27 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr8_+_41386761 | 0.23 |
ENST00000523277.2
|
GINS4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr8_+_24241969 | 0.23 |
ENST00000522298.1
|
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr17_+_47448102 | 0.21 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr1_-_241799232 | 0.21 |
ENST00000366553.1
|
CHML
|
choroideremia-like (Rab escort protein 2) |
| chr8_-_72274095 | 0.20 |
ENST00000303824.7
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr3_-_87325612 | 0.18 |
ENST00000561167.1
ENST00000560656.1 ENST00000344265.3 |
POU1F1
|
POU class 1 homeobox 1 |
| chr8_+_41386725 | 0.16 |
ENST00000276533.3
ENST00000520710.1 ENST00000518671.1 |
GINS4
|
GINS complex subunit 4 (Sld5 homolog) |
| chrX_-_32173579 | 0.14 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr8_+_9413410 | 0.12 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr1_-_120935894 | 0.11 |
ENST00000369383.4
ENST00000369384.4 |
FCGR1B
|
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
| chr6_-_49712147 | 0.08 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr14_-_92198403 | 0.08 |
ENST00000553329.1
ENST00000256343.3 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr11_+_61976137 | 0.05 |
ENST00000244930.4
|
SCGB2A1
|
secretoglobin, family 2A, member 1 |
| chr1_-_205744574 | 0.02 |
ENST00000367139.3
ENST00000235932.4 ENST00000437324.2 ENST00000414729.1 |
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr1_+_196621156 | 0.01 |
ENST00000359637.2
|
CFH
|
complement factor H |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX11
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 14.1 | GO:0051610 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.4 | 4.2 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 1.2 | 7.4 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 1.0 | 3.9 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.9 | 2.8 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.8 | 2.4 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.8 | 9.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.8 | 3.0 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.6 | 3.7 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.5 | 1.6 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.5 | 2.0 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.4 | 2.4 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.4 | 1.6 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 0.4 | 2.3 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.4 | 2.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.3 | 4.6 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.3 | 1.6 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.3 | 1.2 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.3 | 2.7 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.3 | 3.8 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 1.7 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.2 | 3.6 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.2 | 8.2 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.2 | 1.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 1.7 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.2 | 1.3 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.2 | 2.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.2 | 1.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 0.5 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.2 | 7.0 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 1.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.4 | GO:2000354 | response to prolactin(GO:1990637) regulation of ovarian follicle development(GO:2000354) |
| 0.1 | 0.5 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 0.8 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 0.7 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.6 | GO:0035087 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.1 | 5.0 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 1.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.4 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.1 | 1.6 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 1.0 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 1.3 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 2.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.5 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 4.5 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.1 | 0.5 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.5 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 3.9 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 2.3 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.4 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 2.6 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 4.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 5.2 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 1.8 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.8 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 1.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.1 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 2.1 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.4 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.0 | 1.2 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.6 | 2.8 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.5 | 3.8 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.4 | 7.0 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.3 | 1.0 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.3 | 5.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 6.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 4.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.2 | 0.6 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.4 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.1 | 2.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 14.0 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 1.6 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 4.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.4 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 2.2 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 1.7 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 5.3 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.1 | 4.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.4 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 3.7 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.6 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 2.1 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 2.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 4.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 3.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 2.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 3.2 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 1.6 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 1.0 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 6.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 1.3 | 6.7 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.8 | 2.4 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.6 | 1.7 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.6 | 1.7 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.5 | 1.6 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.4 | 1.3 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.4 | 5.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.4 | 1.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.4 | 2.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.3 | 1.7 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.3 | 3.9 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 1.0 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 5.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 0.8 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.2 | 1.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.2 | 0.5 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.2 | 0.6 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.2 | 2.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.2 | 4.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 3.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 2.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 2.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 5.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 4.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 2.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 3.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 4.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 1.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.4 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 7.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 6.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.6 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 10.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.3 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 8.7 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 2.3 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 2.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 4.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 4.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 6.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 6.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 5.0 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 2.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 9.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 4.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 3.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 2.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 2.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.5 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.9 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.3 | 9.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 6.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 8.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 3.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 2.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 2.2 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |