Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
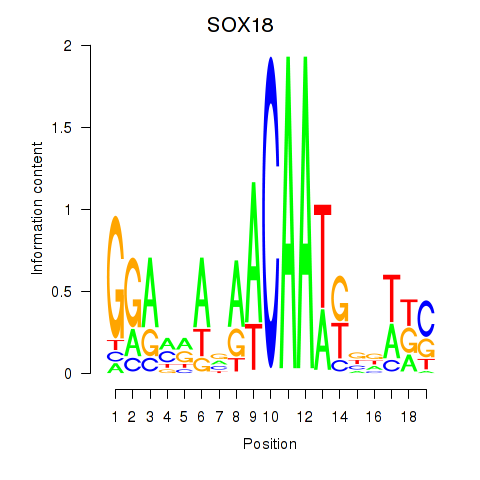
Results for SOX18
Z-value: 0.72
Transcription factors associated with SOX18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX18
|
ENSG00000203883.5 | SRY-box transcription factor 18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX18 | hg19_v2_chr20_-_62680984_62680999 | 0.08 | 2.4e-01 | Click! |
Activity profile of SOX18 motif
Sorted Z-values of SOX18 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_5710919 | 13.52 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chrX_-_70329118 | 12.19 |
ENST00000374188.3
|
IL2RG
|
interleukin 2 receptor, gamma |
| chr4_+_154387480 | 11.37 |
ENST00000409663.3
ENST00000440693.1 ENST00000409959.3 |
KIAA0922
|
KIAA0922 |
| chr3_-_121379739 | 11.12 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr13_-_46756351 | 9.72 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr13_-_46716969 | 9.39 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr2_-_158345462 | 7.40 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chr1_+_209929377 | 6.11 |
ENST00000400959.3
ENST00000367025.3 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr5_+_156607829 | 5.93 |
ENST00000422843.3
|
ITK
|
IL2-inducible T-cell kinase |
| chr11_-_104817919 | 5.43 |
ENST00000533252.1
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr7_-_150329421 | 5.31 |
ENST00000493969.1
ENST00000328902.5 |
GIMAP6
|
GTPase, IMAP family member 6 |
| chr11_-_5248294 | 5.19 |
ENST00000335295.4
|
HBB
|
hemoglobin, beta |
| chr1_+_209929494 | 4.99 |
ENST00000367026.3
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr12_+_25205446 | 4.39 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr21_-_15918618 | 4.36 |
ENST00000400564.1
ENST00000400566.1 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr4_+_109541772 | 4.35 |
ENST00000506397.1
ENST00000394668.2 |
RPL34
|
ribosomal protein L34 |
| chr6_-_26235206 | 4.29 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr6_-_152489484 | 4.19 |
ENST00000354674.4
ENST00000539504.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr6_+_89790490 | 4.11 |
ENST00000336032.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr16_-_68034470 | 4.02 |
ENST00000412757.2
|
DPEP2
|
dipeptidase 2 |
| chr14_-_107219365 | 3.97 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr6_+_89790459 | 3.81 |
ENST00000369472.1
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr13_+_111806083 | 3.77 |
ENST00000375736.4
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr4_+_69962212 | 3.64 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr11_+_71238313 | 3.61 |
ENST00000398536.4
|
KRTAP5-7
|
keratin associated protein 5-7 |
| chr6_-_32908792 | 3.34 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr14_+_22963806 | 3.23 |
ENST00000390493.1
|
TRAJ44
|
T cell receptor alpha joining 44 |
| chr4_+_69962185 | 3.21 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr16_+_33629600 | 3.05 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr6_-_33385854 | 2.99 |
ENST00000488478.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr4_-_57524061 | 2.95 |
ENST00000508121.1
|
HOPX
|
HOP homeobox |
| chr6_-_33385870 | 2.80 |
ENST00000488034.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr6_-_33385655 | 2.75 |
ENST00000440279.3
ENST00000607266.1 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr4_+_109541740 | 2.67 |
ENST00000394665.1
|
RPL34
|
ribosomal protein L34 |
| chr17_+_41150479 | 2.65 |
ENST00000589913.1
|
RPL27
|
ribosomal protein L27 |
| chr4_+_109541722 | 2.63 |
ENST00000394667.3
ENST00000502534.1 |
RPL34
|
ribosomal protein L34 |
| chr17_+_41150290 | 2.58 |
ENST00000589037.1
ENST00000253788.5 |
RPL27
|
ribosomal protein L27 |
| chr1_+_22979474 | 2.55 |
ENST00000509305.1
|
C1QB
|
complement component 1, q subcomponent, B chain |
| chr6_-_33385902 | 2.54 |
ENST00000374500.5
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr1_-_206785898 | 2.43 |
ENST00000271764.2
|
EIF2D
|
eukaryotic translation initiation factor 2D |
| chr17_+_68071389 | 2.43 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr12_-_772901 | 2.41 |
ENST00000305108.4
|
NINJ2
|
ninjurin 2 |
| chr3_+_186435137 | 2.40 |
ENST00000447445.1
|
KNG1
|
kininogen 1 |
| chr2_+_1418154 | 2.40 |
ENST00000423320.1
ENST00000382198.1 |
TPO
|
thyroid peroxidase |
| chr11_-_89224139 | 2.37 |
ENST00000413594.2
|
NOX4
|
NADPH oxidase 4 |
| chr1_-_153517473 | 2.36 |
ENST00000368715.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr5_+_54398463 | 2.36 |
ENST00000274306.6
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
| chr4_+_156588115 | 2.35 |
ENST00000455639.2
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chrX_-_154033661 | 2.30 |
ENST00000393531.1
|
MPP1
|
membrane protein, palmitoylated 1, 55kDa |
| chr6_-_33385823 | 2.28 |
ENST00000494751.1
ENST00000374496.3 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr17_+_68071458 | 2.19 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chrX_+_100645812 | 2.18 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr1_-_206785789 | 2.14 |
ENST00000437518.1
ENST00000367114.3 |
EIF2D
|
eukaryotic translation initiation factor 2D |
| chr19_+_41256764 | 2.14 |
ENST00000243563.3
ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr22_+_37678424 | 2.12 |
ENST00000248901.6
|
CYTH4
|
cytohesin 4 |
| chr2_+_1488435 | 2.09 |
ENST00000446278.1
ENST00000469607.1 |
TPO
|
thyroid peroxidase |
| chr7_-_148725733 | 2.09 |
ENST00000286091.4
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr3_+_108541545 | 2.08 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr18_+_32621324 | 2.08 |
ENST00000300249.5
ENST00000538170.2 ENST00000588910.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr11_-_89224508 | 2.02 |
ENST00000525196.1
|
NOX4
|
NADPH oxidase 4 |
| chr19_+_8117881 | 2.01 |
ENST00000390669.3
|
CCL25
|
chemokine (C-C motif) ligand 25 |
| chr11_-_6677018 | 2.00 |
ENST00000299441.3
|
DCHS1
|
dachsous cadherin-related 1 |
| chr3_+_186330712 | 1.97 |
ENST00000411641.2
ENST00000273784.5 |
AHSG
|
alpha-2-HS-glycoprotein |
| chr10_+_70715884 | 1.95 |
ENST00000354185.4
|
DDX21
|
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr7_+_99933730 | 1.94 |
ENST00000610247.1
|
PILRB
|
paired immunoglobin-like type 2 receptor beta |
| chr15_+_44084040 | 1.93 |
ENST00000249786.4
|
SERF2
|
small EDRK-rich factor 2 |
| chr11_+_64949343 | 1.90 |
ENST00000279247.6
ENST00000532285.1 ENST00000534373.1 |
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr11_+_13299186 | 1.87 |
ENST00000527998.1
ENST00000396441.3 ENST00000533520.1 ENST00000529825.1 ENST00000389707.4 ENST00000401424.1 ENST00000529388.1 ENST00000530357.1 ENST00000403290.1 ENST00000361003.4 ENST00000389708.3 ENST00000403510.3 ENST00000482049.1 |
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like |
| chr6_-_26216872 | 1.82 |
ENST00000244601.3
|
HIST1H2BG
|
histone cluster 1, H2bg |
| chrX_+_47004639 | 1.81 |
ENST00000345781.6
|
RBM10
|
RNA binding motif protein 10 |
| chr4_+_156588249 | 1.76 |
ENST00000393832.3
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr13_-_45992473 | 1.76 |
ENST00000539591.1
ENST00000519676.1 ENST00000519547.1 |
SLC25A30
|
solute carrier family 25, member 30 |
| chr1_-_32801825 | 1.75 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chrX_+_47004599 | 1.73 |
ENST00000329236.7
|
RBM10
|
RNA binding motif protein 10 |
| chr11_-_89224299 | 1.72 |
ENST00000343727.5
ENST00000531342.1 ENST00000375979.3 |
NOX4
|
NADPH oxidase 4 |
| chr8_-_86253888 | 1.64 |
ENST00000522389.1
ENST00000432364.2 ENST00000517618.1 |
CA1
|
carbonic anhydrase I |
| chr2_+_202125219 | 1.59 |
ENST00000323492.7
|
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chrX_-_47004437 | 1.58 |
ENST00000276062.8
|
NDUFB11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
| chr19_+_41257084 | 1.57 |
ENST00000601393.1
|
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr2_+_217277137 | 1.56 |
ENST00000430374.1
ENST00000357276.4 ENST00000444508.1 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr13_-_45992541 | 1.54 |
ENST00000522438.1
|
SLC25A30
|
solute carrier family 25, member 30 |
| chr3_+_186435065 | 1.54 |
ENST00000287611.2
ENST00000265023.4 |
KNG1
|
kininogen 1 |
| chr19_+_8117636 | 1.54 |
ENST00000253451.4
ENST00000315626.4 |
CCL25
|
chemokine (C-C motif) ligand 25 |
| chr15_+_91473403 | 1.48 |
ENST00000394275.2
|
UNC45A
|
unc-45 homolog A (C. elegans) |
| chr3_+_20081515 | 1.47 |
ENST00000263754.4
|
KAT2B
|
K(lysine) acetyltransferase 2B |
| chr1_-_182360918 | 1.47 |
ENST00000339526.4
|
GLUL
|
glutamate-ammonia ligase |
| chr2_+_217277466 | 1.42 |
ENST00000358207.5
ENST00000434435.1 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr1_-_157746909 | 1.41 |
ENST00000392274.3
ENST00000361516.3 ENST00000368181.4 |
FCRL2
|
Fc receptor-like 2 |
| chr12_-_11002063 | 1.38 |
ENST00000544994.1
ENST00000228811.4 ENST00000540107.1 |
PRR4
|
proline rich 4 (lacrimal) |
| chr11_-_89224488 | 1.36 |
ENST00000534731.1
ENST00000527626.1 |
NOX4
|
NADPH oxidase 4 |
| chr12_-_371994 | 1.36 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr2_-_211168332 | 1.34 |
ENST00000341685.4
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr3_+_133465228 | 1.33 |
ENST00000482271.1
ENST00000264998.3 |
TF
|
transferrin |
| chr12_+_96252706 | 1.33 |
ENST00000266735.5
ENST00000553192.1 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr1_-_182361327 | 1.32 |
ENST00000331872.6
ENST00000311223.5 |
GLUL
|
glutamate-ammonia ligase |
| chr12_+_15475462 | 1.30 |
ENST00000543886.1
ENST00000348962.2 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr15_-_68497657 | 1.29 |
ENST00000448060.2
ENST00000467889.1 |
CALML4
|
calmodulin-like 4 |
| chr2_+_79252822 | 1.27 |
ENST00000272324.5
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr19_+_44576296 | 1.27 |
ENST00000421176.3
|
ZNF284
|
zinc finger protein 284 |
| chr6_-_160679905 | 1.27 |
ENST00000366953.3
|
SLC22A2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr12_-_8088871 | 1.24 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr11_+_64949899 | 1.23 |
ENST00000531068.1
ENST00000527699.1 ENST00000533909.1 ENST00000527323.1 |
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr2_-_208031943 | 1.22 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr4_-_144940477 | 1.20 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chr2_+_44502597 | 1.18 |
ENST00000260649.6
ENST00000409387.1 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr22_-_22292934 | 1.14 |
ENST00000538191.1
ENST00000424647.1 ENST00000407142.1 |
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr9_-_123812542 | 1.14 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr4_-_76944621 | 1.07 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr12_-_11422739 | 1.07 |
ENST00000279573.7
|
PRB3
|
proline-rich protein BstNI subfamily 3 |
| chr2_+_90259593 | 1.07 |
ENST00000471857.1
|
IGKV1D-8
|
immunoglobulin kappa variable 1D-8 |
| chr2_+_79252834 | 1.05 |
ENST00000409471.1
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr11_-_89223883 | 1.04 |
ENST00000528341.1
|
NOX4
|
NADPH oxidase 4 |
| chr9_-_127533519 | 1.03 |
ENST00000487099.2
ENST00000344523.4 ENST00000373584.3 |
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr1_+_66258846 | 1.03 |
ENST00000341517.4
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr16_-_787771 | 1.03 |
ENST00000568545.1
|
NARFL
|
nuclear prelamin A recognition factor-like |
| chrX_+_55744228 | 1.03 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
| chr19_+_33571786 | 1.00 |
ENST00000170564.2
|
GPATCH1
|
G patch domain containing 1 |
| chr1_+_171154347 | 1.00 |
ENST00000209929.7
ENST00000441535.1 |
FMO2
|
flavin containing monooxygenase 2 (non-functional) |
| chr15_+_64386261 | 0.99 |
ENST00000560829.1
|
SNX1
|
sorting nexin 1 |
| chr12_+_113344755 | 0.98 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr17_+_56315936 | 0.96 |
ENST00000543544.1
|
LPO
|
lactoperoxidase |
| chr1_+_13521973 | 0.96 |
ENST00000327795.5
|
PRAMEF21
|
PRAME family member 21 |
| chr1_+_36348790 | 0.91 |
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr11_+_134201911 | 0.88 |
ENST00000389881.3
|
GLB1L2
|
galactosidase, beta 1-like 2 |
| chr2_+_79252804 | 0.87 |
ENST00000393897.2
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr5_+_32711419 | 0.85 |
ENST00000265074.8
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr11_-_118047376 | 0.83 |
ENST00000278947.5
|
SCN2B
|
sodium channel, voltage-gated, type II, beta subunit |
| chr17_+_3118915 | 0.82 |
ENST00000304094.1
|
OR1A1
|
olfactory receptor, family 1, subfamily A, member 1 |
| chrX_-_101726732 | 0.82 |
ENST00000457521.2
ENST00000412230.2 ENST00000453326.2 |
NXF2B
TCP11X2
|
nuclear RNA export factor 2B t-complex 11 family, X-linked 2 |
| chr5_-_179050066 | 0.82 |
ENST00000329433.6
ENST00000510411.1 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr10_-_105992059 | 0.80 |
ENST00000369720.1
ENST00000369719.1 ENST00000357060.3 ENST00000428666.1 ENST00000278064.2 |
WDR96
|
WD repeat domain 96 |
| chr1_-_12958101 | 0.80 |
ENST00000235347.4
|
PRAMEF10
|
PRAME family member 10 |
| chr9_-_99064386 | 0.79 |
ENST00000375262.2
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr8_+_40010989 | 0.78 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr11_+_20620946 | 0.76 |
ENST00000525748.1
|
SLC6A5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr8_-_37707356 | 0.76 |
ENST00000520601.1
ENST00000521170.1 ENST00000220659.6 |
BRF2
|
BRF2, RNA polymerase III transcription initiation factor 50 kDa subunit |
| chr9_-_74525658 | 0.74 |
ENST00000333421.6
|
ABHD17B
|
abhydrolase domain containing 17B |
| chr12_-_10562356 | 0.73 |
ENST00000309384.1
|
KLRC4
|
killer cell lectin-like receptor subfamily C, member 4 |
| chr1_-_156828810 | 0.73 |
ENST00000368195.3
|
INSRR
|
insulin receptor-related receptor |
| chr11_+_74699942 | 0.70 |
ENST00000526068.1
ENST00000532963.1 ENST00000531619.1 ENST00000534628.1 ENST00000545272.1 |
NEU3
|
sialidase 3 (membrane sialidase) |
| chrX_+_101470280 | 0.69 |
ENST00000395088.2
ENST00000330252.5 ENST00000333110.5 |
NXF2
TCP11X1
|
nuclear RNA export factor 2 t-complex 11 family, X-linked 1 |
| chr12_-_110939870 | 0.68 |
ENST00000447578.2
ENST00000546588.1 ENST00000360579.7 ENST00000549970.1 ENST00000549578.1 |
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr9_-_74525847 | 0.65 |
ENST00000377041.2
|
ABHD17B
|
abhydrolase domain containing 17B |
| chr12_-_68647281 | 0.61 |
ENST00000328087.4
ENST00000538666.1 |
IL22
|
interleukin 22 |
| chr6_+_26217159 | 0.61 |
ENST00000303910.2
|
HIST1H2AE
|
histone cluster 1, H2ae |
| chr17_+_72427477 | 0.60 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr16_+_28770025 | 0.59 |
ENST00000435324.2
|
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr5_+_140800638 | 0.59 |
ENST00000398587.2
ENST00000518882.1 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chrX_+_55744166 | 0.59 |
ENST00000374941.4
ENST00000414239.1 |
RRAGB
|
Ras-related GTP binding B |
| chr3_-_37034702 | 0.59 |
ENST00000322716.5
|
EPM2AIP1
|
EPM2A (laforin) interacting protein 1 |
| chr5_-_75919217 | 0.58 |
ENST00000504899.1
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr8_+_38644778 | 0.57 |
ENST00000276520.8
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr9_-_21351377 | 0.57 |
ENST00000380210.1
|
IFNA6
|
interferon, alpha 6 |
| chr19_+_36132631 | 0.55 |
ENST00000379026.2
ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2
|
ets variant 2 |
| chr6_-_133035185 | 0.55 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr1_+_160051319 | 0.54 |
ENST00000368088.3
|
KCNJ9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr11_-_75236867 | 0.53 |
ENST00000376282.3
ENST00000336898.3 |
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr3_-_197300194 | 0.53 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chrX_+_119495934 | 0.51 |
ENST00000218008.3
ENST00000361319.3 ENST00000539306.1 |
ATP1B4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide |
| chr14_+_24458123 | 0.48 |
ENST00000545240.1
ENST00000382755.4 |
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4 like 2 |
| chr9_+_33240157 | 0.46 |
ENST00000379721.3
|
SPINK4
|
serine peptidase inhibitor, Kazal type 4 |
| chr7_-_6048702 | 0.45 |
ENST00000265849.7
|
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr8_-_102218292 | 0.41 |
ENST00000518336.1
ENST00000520454.1 |
ZNF706
|
zinc finger protein 706 |
| chr10_-_127505167 | 0.41 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr11_+_60970852 | 0.39 |
ENST00000325558.6
|
PGA3
|
pepsinogen 3, group I (pepsinogen A) |
| chr2_-_39347524 | 0.36 |
ENST00000395038.2
ENST00000402219.2 |
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr1_+_153003671 | 0.35 |
ENST00000307098.4
|
SPRR1B
|
small proline-rich protein 1B |
| chr17_+_25799008 | 0.35 |
ENST00000583370.1
ENST00000398988.3 ENST00000268763.6 |
KSR1
|
kinase suppressor of ras 1 |
| chr6_+_42883727 | 0.34 |
ENST00000304672.1
ENST00000441198.1 ENST00000446507.1 |
PTCRA
|
pre T-cell antigen receptor alpha |
| chr11_-_76155618 | 0.34 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr4_-_39367949 | 0.34 |
ENST00000503784.1
ENST00000349703.2 ENST00000381897.1 |
RFC1
|
replication factor C (activator 1) 1, 145kDa |
| chrX_+_2984874 | 0.31 |
ENST00000359361.2
|
ARSF
|
arylsulfatase F |
| chr12_-_48963829 | 0.31 |
ENST00000301046.2
ENST00000549817.1 |
LALBA
|
lactalbumin, alpha- |
| chr9_+_117350009 | 0.29 |
ENST00000374050.3
|
ATP6V1G1
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1 |
| chr5_+_135468516 | 0.29 |
ENST00000507118.1
ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5
|
SMAD family member 5 |
| chr3_-_184971853 | 0.28 |
ENST00000231887.3
|
EHHADH
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr7_-_142176790 | 0.27 |
ENST00000390369.2
|
TRBV7-4
|
T cell receptor beta variable 7-4 (gene/pseudogene) |
| chr1_+_206223941 | 0.27 |
ENST00000367126.4
|
AVPR1B
|
arginine vasopressin receptor 1B |
| chr17_+_48503519 | 0.27 |
ENST00000300441.4
ENST00000541920.1 ENST00000506582.1 ENST00000504392.1 ENST00000427954.2 |
ACSF2
|
acyl-CoA synthetase family member 2 |
| chr2_+_105953972 | 0.26 |
ENST00000410049.1
|
C2orf49
|
chromosome 2 open reading frame 49 |
| chr6_+_29427548 | 0.25 |
ENST00000377132.1
|
OR2H1
|
olfactory receptor, family 2, subfamily H, member 1 |
| chr17_+_48503603 | 0.23 |
ENST00000502667.1
|
ACSF2
|
acyl-CoA synthetase family member 2 |
| chrX_-_2847366 | 0.22 |
ENST00000381154.1
|
ARSD
|
arylsulfatase D |
| chr2_+_241544834 | 0.22 |
ENST00000319838.5
ENST00000403859.1 ENST00000438013.2 |
GPR35
|
G protein-coupled receptor 35 |
| chr4_+_54243798 | 0.21 |
ENST00000337488.6
ENST00000358575.5 ENST00000507922.1 |
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr7_-_6048650 | 0.21 |
ENST00000382321.4
ENST00000406569.3 |
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr1_-_241799232 | 0.21 |
ENST00000366553.1
|
CHML
|
choroideremia-like (Rab escort protein 2) |
| chr4_+_164265035 | 0.21 |
ENST00000338566.3
|
NPY5R
|
neuropeptide Y receptor Y5 |
| chr12_+_69633317 | 0.21 |
ENST00000435070.2
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr9_-_99064429 | 0.20 |
ENST00000375263.3
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr2_-_49381646 | 0.20 |
ENST00000346173.3
ENST00000406846.2 |
FSHR
|
follicle stimulating hormone receptor |
| chr19_-_57347415 | 0.20 |
ENST00000601070.1
|
ZIM2
|
zinc finger, imprinted 2 |
| chr5_+_32711829 | 0.19 |
ENST00000415167.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr13_-_99630233 | 0.18 |
ENST00000376460.1
ENST00000442173.1 |
DOCK9
|
dedicator of cytokinesis 9 |
| chr2_-_49381572 | 0.18 |
ENST00000454032.1
ENST00000304421.4 |
FSHR
|
follicle stimulating hormone receptor |
| chr9_+_74526384 | 0.18 |
ENST00000334731.2
ENST00000377031.3 |
C9orf85
|
chromosome 9 open reading frame 85 |
| chr12_+_113344582 | 0.17 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr19_-_46105411 | 0.14 |
ENST00000323040.4
ENST00000544371.1 |
GPR4
OPA3
|
G protein-coupled receptor 4 optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr1_+_100316041 | 0.13 |
ENST00000370165.3
ENST00000370163.3 ENST00000294724.4 |
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr9_+_33629119 | 0.12 |
ENST00000331828.4
|
TRBV21OR9-2
|
T cell receptor beta variable 21/OR9-2 (pseudogene) |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX18
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 12.2 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 1.3 | 5.2 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 1.2 | 19.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 1.1 | 3.3 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 1.1 | 3.2 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 1.0 | 7.9 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.7 | 12.7 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.7 | 3.5 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.7 | 2.8 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.7 | 4.1 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.7 | 2.0 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.6 | 5.9 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.5 | 4.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.5 | 4.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.5 | 5.4 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.5 | 1.5 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.5 | 1.9 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.4 | 1.3 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.4 | 4.2 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.4 | 1.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.4 | 1.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.4 | 3.8 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.3 | 0.3 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.3 | 3.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.3 | 2.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 13.5 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.2 | 3.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.2 | 8.5 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.2 | 2.4 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.2 | 1.6 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.2 | 5.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 1.3 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.2 | 1.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.2 | 1.0 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.2 | 1.2 | GO:1990822 | L-cystine transport(GO:0015811) basic amino acid transmembrane transport(GO:1990822) |
| 0.2 | 0.9 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.2 | 4.5 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.2 | 3.9 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.2 | 3.7 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.1 | 0.8 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 3.0 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 0.8 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 | 1.3 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.1 | 1.0 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 1.1 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 1.0 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 1.1 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 | 0.6 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 5.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 4.4 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 17.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 2.1 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.5 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.4 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 0.1 | 7.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.8 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 0.4 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.1 | 1.0 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.1 | 1.9 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 2.0 | GO:0030502 | pinocytosis(GO:0006907) negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 0.7 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 1.0 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 2.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.6 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 1.4 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 | 0.4 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.1 | 1.0 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 2.3 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.1 | 0.6 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 0.4 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 3.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.7 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 0.5 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.1 | 0.2 | GO:0002865 | negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) |
| 0.0 | 2.5 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.3 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 1.3 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 1.4 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.3 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.7 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 4.4 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 1.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.7 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.7 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.8 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 1.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 9.0 | GO:0010038 | response to metal ion(GO:0010038) |
| 0.0 | 0.3 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 1.3 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.1 | GO:0072144 | renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 2.5 | GO:0048839 | inner ear development(GO:0048839) |
| 0.0 | 0.3 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.4 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.1 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.1 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 2.4 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 1.1 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.8 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.5 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 1.4 | GO:0031424 | keratinization(GO:0031424) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.0 | 19.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.9 | 5.4 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.5 | 2.5 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.5 | 8.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.5 | 3.8 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.3 | 4.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.3 | 1.6 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.3 | 1.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 0.7 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.2 | 1.0 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.2 | 3.0 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 12.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 1.9 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 1.3 | GO:0034715 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.2 | 4.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 2.8 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.2 | 1.5 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.2 | 17.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 3.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.0 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 7.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.7 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 1.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 4.3 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 0.9 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 0.7 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 1.6 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 3.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 2.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 2.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 4.9 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 7.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 4.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 2.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 12.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.4 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 1.0 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 4.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 1.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.5 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 0.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 1.1 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 8.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 7.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 9.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.3 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 1.2 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 1.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 3.2 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 1.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.1 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.2 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) |
| 0.9 | 3.5 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.9 | 5.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.9 | 8.5 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.7 | 4.5 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.7 | 2.8 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.5 | 3.7 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.4 | 1.3 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.4 | 1.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.4 | 5.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.3 | 1.6 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.3 | 1.3 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.3 | 3.0 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.3 | 3.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.3 | 5.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 1.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 1.9 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 1.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 1.4 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.2 | 1.6 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 1.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.2 | 1.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.2 | 0.8 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.2 | 4.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 4.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 0.5 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.2 | 0.7 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.2 | 1.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 0.8 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.2 | 1.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.2 | 4.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 12.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.2 | 0.8 | GO:0001016 | transcription factor activity, RNA polymerase III transcription factor binding(GO:0001007) RNA polymerase III regulatory region DNA binding(GO:0001016) |
| 0.1 | 2.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 5.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.9 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.5 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 7.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 1.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.6 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 2.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.9 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 3.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 2.1 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.7 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.1 | 0.4 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.1 | 5.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 4.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 2.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 3.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.3 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.7 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.3 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.1 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 10.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 14.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 2.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 4.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 12.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.5 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.9 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 3.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 10.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 3.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 1.1 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 4.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 1.0 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 4.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.0 | GO:0019210 | kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 1.0 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.1 | GO:0003823 | antigen binding(GO:0003823) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 12.5 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.2 | 17.0 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.2 | 11.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 3.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 4.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 7.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 2.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 2.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 2.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 11.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 2.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 12.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 2.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 4.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 5.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 3.9 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 4.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 5.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 7.0 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 1.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 2.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 5.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 17.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 2.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.0 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 4.1 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 4.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 6.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 5.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 3.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.8 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 3.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.6 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |