Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
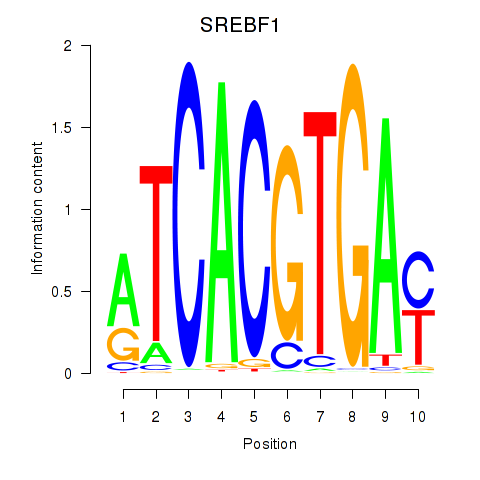
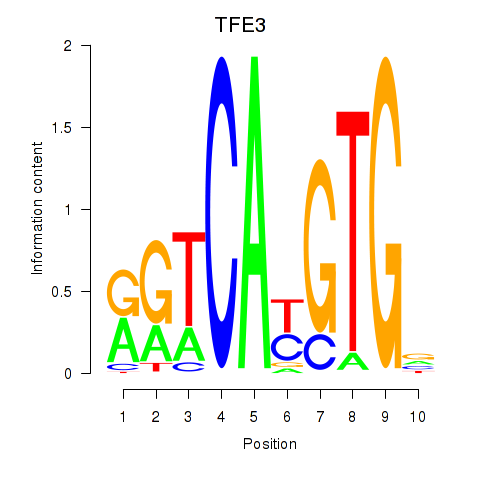
Results for SREBF1_TFE3
Z-value: 1.39
Transcription factors associated with SREBF1_TFE3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SREBF1
|
ENSG00000072310.12 | sterol regulatory element binding transcription factor 1 |
|
TFE3
|
ENSG00000068323.12 | transcription factor binding to IGHM enhancer 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFE3 | hg19_v2_chrX_-_48901012_48901050 | 0.43 | 2.2e-11 | Click! |
| SREBF1 | hg19_v2_chr17_-_17740287_17740316 | -0.08 | 2.4e-01 | Click! |
Activity profile of SREBF1_TFE3 motif
Sorted Z-values of SREBF1_TFE3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SREBF1_TFE3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 20.3 | 81.2 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 15.2 | 60.9 | GO:0002086 | maltose metabolic process(GO:0000023) diaphragm contraction(GO:0002086) |
| 12.3 | 61.3 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 6.9 | 41.5 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 6.7 | 20.2 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 6.3 | 18.9 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 5.5 | 49.4 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 5.3 | 58.8 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 5.1 | 20.6 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 4.8 | 14.4 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 4.6 | 13.7 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 4.4 | 17.5 | GO:1900241 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 4.3 | 13.0 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 4.3 | 21.4 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 4.1 | 24.4 | GO:0015811 | L-cystine transport(GO:0015811) |
| 4.0 | 20.1 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 3.9 | 27.5 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 3.9 | 31.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 3.6 | 10.9 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 3.6 | 18.1 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 3.6 | 10.7 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 3.5 | 74.4 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 3.4 | 34.3 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 3.4 | 16.9 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 3.3 | 9.9 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 3.2 | 16.1 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 3.0 | 9.1 | GO:0051039 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 3.0 | 9.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 2.8 | 53.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 2.8 | 5.6 | GO:0048286 | lung alveolus development(GO:0048286) |
| 2.7 | 2.7 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 2.7 | 10.7 | GO:0061528 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 2.6 | 12.9 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 2.6 | 10.2 | GO:0006726 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 2.4 | 21.9 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 2.3 | 7.0 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 2.3 | 9.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 2.2 | 8.9 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 2.2 | 4.4 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 2.2 | 17.4 | GO:0070459 | prolactin secretion(GO:0070459) |
| 2.1 | 10.6 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 2.0 | 6.1 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 2.0 | 56.6 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 2.0 | 6.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 2.0 | 11.8 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 2.0 | 11.8 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 2.0 | 33.2 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 1.9 | 7.8 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 1.9 | 5.8 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 1.9 | 19.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 1.9 | 15.4 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 1.8 | 12.9 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 1.8 | 5.5 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 1.8 | 5.5 | GO:0002904 | B cell negative selection(GO:0002352) positive regulation of B cell apoptotic process(GO:0002904) post-embryonic camera-type eye morphogenesis(GO:0048597) apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) positive regulation of apoptotic DNA fragmentation(GO:1902512) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 1.8 | 5.4 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 1.8 | 5.4 | GO:0006788 | heme oxidation(GO:0006788) smooth muscle hyperplasia(GO:0014806) |
| 1.8 | 3.6 | GO:1904978 | regulation of endosome organization(GO:1904978) |
| 1.8 | 5.3 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 1.7 | 5.2 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 1.7 | 12.0 | GO:0046465 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 1.7 | 5.0 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 1.7 | 5.0 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 1.7 | 16.8 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 1.6 | 1.6 | GO:1903487 | regulation of lactation(GO:1903487) |
| 1.6 | 9.4 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 1.6 | 6.2 | GO:0023021 | glucosylceramide catabolic process(GO:0006680) termination of signal transduction(GO:0023021) beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 1.6 | 7.8 | GO:1904579 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) response to hypobaric hypoxia(GO:1990910) |
| 1.5 | 4.5 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 1.5 | 15.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 1.5 | 8.9 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 1.5 | 5.9 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 1.5 | 5.9 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 1.5 | 4.4 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 1.4 | 5.7 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 1.4 | 24.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 1.4 | 12.7 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 1.4 | 8.5 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 1.4 | 16.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.3 | 6.7 | GO:1900920 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 1.3 | 7.7 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 1.3 | 20.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 1.3 | 2.5 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 1.2 | 9.9 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 1.2 | 3.7 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 1.2 | 4.8 | GO:0009386 | translational attenuation(GO:0009386) |
| 1.2 | 6.0 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 1.2 | 1.2 | GO:0048754 | branching morphogenesis of an epithelial tube(GO:0048754) morphogenesis of a branching epithelium(GO:0061138) |
| 1.2 | 3.5 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 1.2 | 7.0 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 1.2 | 2.3 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 1.2 | 5.8 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 1.1 | 3.4 | GO:0003285 | septum secundum development(GO:0003285) |
| 1.1 | 24.7 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 1.1 | 4.5 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 1.1 | 5.6 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 1.1 | 13.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 1.1 | 19.0 | GO:0006265 | DNA topological change(GO:0006265) |
| 1.1 | 4.5 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 1.1 | 11.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 1.1 | 3.2 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) circadian temperature homeostasis(GO:0060086) |
| 1.1 | 6.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 1.1 | 3.2 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 1.0 | 1.0 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 1.0 | 3.1 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 1.0 | 11.5 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 1.0 | 4.2 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 1.0 | 3.1 | GO:0071359 | cellular response to dsRNA(GO:0071359) |
| 1.0 | 3.1 | GO:0061055 | myotome development(GO:0061055) |
| 1.0 | 5.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 1.0 | 23.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 1.0 | 12.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 1.0 | 10.0 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 1.0 | 5.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 1.0 | 8.0 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 1.0 | 5.0 | GO:0097338 | response to clozapine(GO:0097338) |
| 1.0 | 29.0 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 1.0 | 6.7 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 1.0 | 7.7 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 1.0 | 1.9 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.9 | 5.6 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.9 | 2.7 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.9 | 22.5 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.9 | 3.6 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.9 | 1.7 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.8 | 10.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.8 | 5.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.8 | 2.5 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.8 | 1.6 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.8 | 18.0 | GO:0007567 | parturition(GO:0007567) |
| 0.8 | 7.4 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.8 | 12.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.8 | 3.2 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.8 | 2.4 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.8 | 4.7 | GO:0060592 | mammary gland formation(GO:0060592) |
| 0.8 | 9.4 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.8 | 2.3 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.8 | 3.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.8 | 5.4 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.8 | 3.0 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.8 | 25.1 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.8 | 2.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.8 | 3.8 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.8 | 10.5 | GO:0035878 | nail development(GO:0035878) |
| 0.7 | 2.2 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.7 | 2.2 | GO:1903012 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) positive regulation of bone mineralization involved in bone maturation(GO:1900159) positive regulation of bone development(GO:1903012) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 0.7 | 6.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.7 | 6.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.7 | 3.5 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.7 | 12.0 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.7 | 4.9 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.7 | 3.5 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.7 | 5.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.7 | 5.5 | GO:0015886 | heme transport(GO:0015886) |
| 0.7 | 8.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.7 | 2.0 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.7 | 2.7 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.7 | 2.0 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.7 | 0.7 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.6 | 3.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.6 | 7.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.6 | 1.9 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.6 | 14.1 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.6 | 7.0 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.6 | 1.9 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.6 | 12.7 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) |
| 0.6 | 7.0 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.6 | 20.2 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.6 | 11.8 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.6 | 3.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.6 | 6.7 | GO:1902224 | ketone body metabolic process(GO:1902224) |
| 0.6 | 3.1 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.6 | 1.8 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.6 | 4.8 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.6 | 15.4 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.6 | 1.8 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.6 | 1.7 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.6 | 1.7 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.6 | 1.7 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.6 | 2.8 | GO:0046618 | drug export(GO:0046618) |
| 0.6 | 14.0 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.6 | 2.2 | GO:0005985 | sucrose metabolic process(GO:0005985) |
| 0.5 | 1.6 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.5 | 15.7 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.5 | 4.3 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.5 | 2.7 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.5 | 3.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.5 | 2.7 | GO:0032898 | nerve growth factor processing(GO:0032455) neurotrophin production(GO:0032898) |
| 0.5 | 2.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.5 | 7.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.5 | 2.5 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.5 | 2.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.5 | 6.5 | GO:0019374 | galactosylceramide metabolic process(GO:0006681) galactolipid metabolic process(GO:0019374) |
| 0.5 | 1.5 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.5 | 1.0 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.5 | 1.5 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.5 | 2.0 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.5 | 5.9 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.5 | 11.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.5 | 5.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.5 | 3.8 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.5 | 3.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) regulation of adipose tissue development(GO:1904177) |
| 0.5 | 5.2 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.5 | 2.8 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.5 | 12.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.5 | 3.2 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.5 | 1.4 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.4 | 4.4 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.4 | 1.7 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.4 | 1.3 | GO:0010887 | detection of endogenous stimulus(GO:0009726) negative regulation of cholesterol storage(GO:0010887) response to high density lipoprotein particle(GO:0055099) |
| 0.4 | 17.6 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.4 | 4.7 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.4 | 57.8 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.4 | 21.2 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.4 | 4.6 | GO:0035646 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.4 | 6.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.4 | 0.8 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.4 | 3.3 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.4 | 3.7 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.4 | 13.9 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.4 | 1.6 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.4 | 4.4 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.4 | 14.3 | GO:1904031 | positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.4 | 1.6 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.4 | 5.8 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.4 | 1.2 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.4 | 2.7 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.4 | 1.1 | GO:1900075 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.4 | 3.0 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.4 | 8.2 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.4 | 1.8 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.4 | 1.1 | GO:1903401 | L-lysine transmembrane transport(GO:1903401) |
| 0.4 | 5.8 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.4 | 24.9 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.4 | 4.6 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.4 | 1.8 | GO:0072021 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.4 | 2.8 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.4 | 1.1 | GO:0001928 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.4 | 9.1 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.3 | 0.3 | GO:0043317 | regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 0.3 | 2.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.3 | 19.5 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.3 | 1.0 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.3 | 2.4 | GO:0090647 | modulation of age-related behavioral decline(GO:0090647) |
| 0.3 | 1.7 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.3 | 1.0 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.3 | 4.7 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.3 | 1.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.3 | 1.0 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.3 | 11.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.3 | 12.7 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.3 | 4.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.3 | 1.6 | GO:1903525 | regulation of membrane tubulation(GO:1903525) positive regulation of membrane tubulation(GO:1903527) |
| 0.3 | 4.5 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.3 | 3.5 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.3 | 9.5 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.3 | 5.3 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.3 | 3.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.3 | 1.6 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.3 | 8.4 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.3 | 25.8 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.3 | 3.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.3 | 0.6 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.3 | 6.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.3 | 8.4 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.3 | 1.4 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.3 | 1.1 | GO:0030879 | mammary gland development(GO:0030879) |
| 0.3 | 2.0 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.3 | 7.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.3 | 4.2 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.3 | 1.1 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.3 | 5.7 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.3 | 1.9 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.3 | 2.2 | GO:0032106 | positive regulation of macroautophagy(GO:0016239) positive regulation of response to extracellular stimulus(GO:0032106) positive regulation of response to nutrient levels(GO:0032109) |
| 0.3 | 0.8 | GO:0042823 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.3 | 1.1 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.3 | 3.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.3 | 1.0 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.3 | 1.0 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.2 | 2.0 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.2 | 2.5 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.2 | 1.0 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.2 | 8.9 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.2 | 3.3 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.2 | 0.7 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.2 | 2.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 | 0.7 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.2 | 1.6 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.2 | 1.4 | GO:0061525 | hindgut morphogenesis(GO:0007442) embryonic hindgut morphogenesis(GO:0048619) hindgut development(GO:0061525) |
| 0.2 | 3.6 | GO:0019941 | modification-dependent protein catabolic process(GO:0019941) modification-dependent macromolecule catabolic process(GO:0043632) |
| 0.2 | 12.5 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.2 | 2.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 7.1 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.2 | 6.0 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.2 | 0.6 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.2 | 1.9 | GO:0042423 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.2 | 6.1 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.2 | 1.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 1.8 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.2 | 2.0 | GO:0045329 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.2 | 1.0 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.2 | 2.4 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 2.4 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.2 | 3.0 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 3.8 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 1.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 1.6 | GO:0021527 | ventral spinal cord interneuron differentiation(GO:0021514) spinal cord association neuron differentiation(GO:0021527) |
| 0.2 | 0.8 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.2 | 4.1 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.2 | 1.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 0.4 | GO:1903726 | negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.2 | 3.8 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.2 | 3.0 | GO:0097503 | sialylation(GO:0097503) |
| 0.2 | 3.2 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.2 | 0.9 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.2 | 1.3 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.2 | 25.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.2 | 0.5 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.2 | 8.3 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.2 | 1.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 2.5 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.2 | 12.8 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.2 | 1.6 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.2 | 3.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 0.7 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.2 | 0.7 | GO:0071205 | protein localization to paranode region of axon(GO:0002175) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.2 | 2.5 | GO:0021895 | cerebral cortex neuron differentiation(GO:0021895) |
| 0.2 | 4.0 | GO:0007530 | sex determination(GO:0007530) |
| 0.2 | 1.2 | GO:0051603 | proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.2 | 1.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 0.5 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.2 | 1.3 | GO:0007270 | neuron-neuron synaptic transmission(GO:0007270) |
| 0.2 | 2.5 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 1.7 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.2 | 0.6 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.2 | 5.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.2 | 4.9 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.2 | 4.9 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.2 | 0.8 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 1.5 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.2 | 8.9 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.6 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.1 | 0.7 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 5.3 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 1.0 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 1.6 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 2.4 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 1.2 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.5 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 1.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 3.5 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 0.6 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 1.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 3.5 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.1 | 6.0 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 5.8 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.1 | 2.1 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 0.6 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.1 | 1.4 | GO:0036508 | protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.1 | 0.4 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 1.9 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 0.8 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.1 | 0.4 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 | 1.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 2.4 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 1.4 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.7 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 0.5 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 0.3 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.4 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.1 | 0.3 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 1.5 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.1 | 5.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.4 | GO:0044241 | lipid digestion(GO:0044241) |
| 0.1 | 4.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.7 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.1 | 6.1 | GO:0006023 | aminoglycan biosynthetic process(GO:0006023) glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.1 | 0.3 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.1 | 1.3 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.1 | 2.6 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.1 | 0.5 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 1.1 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 0.7 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 15.3 | GO:0006457 | protein folding(GO:0006457) |
| 0.1 | 2.0 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 1.7 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 | 1.4 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.1 | 0.4 | GO:0045359 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.1 | 0.5 | GO:0097104 | postsynaptic membrane assembly(GO:0097104) |
| 0.1 | 5.8 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.1 | 0.4 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 3.0 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 0.1 | GO:1902954 | early endosome to recycling endosome transport(GO:0061502) regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.1 | 0.9 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.1 | 8.0 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.1 | 0.9 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.1 | 2.2 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.1 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 1.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.2 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.1 | 1.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 2.9 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.1 | 2.9 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 6.5 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 2.0 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 1.8 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 0.6 | GO:1903432 | regulation of TORC1 signaling(GO:1903432) |
| 0.1 | 0.2 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 | 0.5 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.1 | 0.8 | GO:0046320 | regulation of fatty acid oxidation(GO:0046320) |
| 0.1 | 1.8 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 4.2 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.3 | GO:0070875 | positive regulation of glycogen biosynthetic process(GO:0045725) positive regulation of glycogen metabolic process(GO:0070875) |
| 0.0 | 0.5 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 0.7 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 2.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.4 | GO:1901032 | negative regulation of response to reactive oxygen species(GO:1901032) negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.0 | 0.4 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.5 | GO:1900116 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 1.7 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 1.5 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 1.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 1.0 | GO:0016311 | dephosphorylation(GO:0016311) |
| 0.0 | 1.0 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.0 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 1.3 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 1.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 2.2 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.5 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.5 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.5 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 3.1 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 1.3 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 1.1 | GO:0032945 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) negative regulation of leukocyte proliferation(GO:0070664) |
| 0.0 | 0.6 | GO:1903169 | regulation of calcium ion transmembrane transport(GO:1903169) |
| 0.0 | 0.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.5 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.1 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.3 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.3 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.3 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.1 | 96.5 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 10.4 | 41.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 7.7 | 53.6 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 6.4 | 32.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 6.3 | 50.4 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 4.3 | 17.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 4.2 | 58.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 3.9 | 15.8 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 3.4 | 10.1 | GO:0019034 | viral replication complex(GO:0019034) |
| 2.7 | 37.6 | GO:0030897 | HOPS complex(GO:0030897) |
| 2.7 | 10.7 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 2.5 | 25.4 | GO:0097413 | Lewy body(GO:0097413) |
| 2.5 | 14.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 2.3 | 13.5 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 2.2 | 8.8 | GO:0033263 | CORVET complex(GO:0033263) |
| 2.1 | 15.0 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 1.8 | 21.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 1.7 | 245.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 1.6 | 6.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 1.5 | 10.8 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 1.5 | 6.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 1.5 | 11.9 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 1.4 | 9.8 | GO:0097452 | GAIT complex(GO:0097452) |
| 1.2 | 6.0 | GO:0089701 | U2AF(GO:0089701) |
| 1.1 | 5.6 | GO:0071546 | perinucleolar chromocenter(GO:0010370) pi-body(GO:0071546) |
| 1.1 | 18.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 1.1 | 7.7 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 1.1 | 16.9 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 1.0 | 23.2 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.9 | 50.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.9 | 6.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.9 | 14.2 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.8 | 2.5 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.8 | 6.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.7 | 5.9 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.7 | 3.3 | GO:0000801 | central element(GO:0000801) |
| 0.7 | 4.0 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.6 | 15.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.6 | 16.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.6 | 13.5 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.6 | 8.8 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.5 | 43.9 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.5 | 1.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.5 | 1.0 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.5 | 4.6 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.5 | 3.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.5 | 5.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.5 | 5.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.5 | 4.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.5 | 3.7 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.5 | 6.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.4 | 7.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.4 | 3.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.4 | 4.7 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.4 | 1.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.4 | 3.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.4 | 19.7 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.4 | 38.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.4 | 18.9 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.4 | 5.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.4 | 50.7 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.4 | 6.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.4 | 33.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.4 | 8.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.4 | 2.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.3 | 1.7 | GO:0033503 | HULC complex(GO:0033503) |
| 0.3 | 4.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.3 | 1.3 | GO:0002177 | manchette(GO:0002177) |
| 0.3 | 8.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.3 | 9.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.3 | 3.1 | GO:0070938 | contractile ring(GO:0070938) |
| 0.3 | 20.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.3 | 23.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.3 | 1.9 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.3 | 2.2 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.3 | 12.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 12.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.3 | 3.6 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.3 | 2.0 | GO:0032059 | bleb(GO:0032059) |
| 0.3 | 1.7 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.3 | 4.3 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.3 | 3.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.3 | 6.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.3 | 2.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.3 | 1.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.3 | 4.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.3 | 8.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.3 | 82.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.3 | 3.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 44.9 | GO:0005770 | late endosome(GO:0005770) |
| 0.3 | 6.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.3 | 6.7 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.3 | 39.6 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.3 | 3.3 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.2 | 5.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 17.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.2 | 11.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 16.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 0.7 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 2.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 3.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.2 | 16.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.2 | 5.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 16.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 2.0 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.2 | 4.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 1.7 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.2 | 0.6 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.2 | 12.4 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.2 | 20.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 1.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 9.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 3.7 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.2 | 1.1 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.2 | 1.9 | GO:0042611 | MHC protein complex(GO:0042611) MHC class I protein complex(GO:0042612) |
| 0.2 | 9.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 10.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 8.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 18.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.2 | 4.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 8.0 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 2.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 16.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 21.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 7.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 2.0 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 1.9 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 13.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 1.9 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 3.6 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 26.6 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 3.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 4.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 95.4 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 10.2 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 3.0 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 11.4 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.1 | 1.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 27.4 | GO:0005929 | cilium(GO:0005929) |
| 0.1 | 0.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 2.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 4.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 103.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 0.5 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 1.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 1.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 2.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 2.1 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.2 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 1.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.6 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 4.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 2.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 3.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.0 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 2.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 70.0 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0045277 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 20.3 | 60.9 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 12.3 | 61.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 11.3 | 45.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 7.1 | 28.3 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 7.0 | 56.4 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 5.0 | 15.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 4.8 | 19.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 4.8 | 14.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 4.7 | 18.9 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 4.4 | 26.6 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 4.3 | 12.8 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 4.1 | 24.4 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 4.0 | 23.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 3.9 | 31.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 3.6 | 10.9 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 3.6 | 39.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 3.5 | 10.4 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 3.5 | 31.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 3.3 | 73.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 3.2 | 12.9 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 3.2 | 73.8 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 3.2 | 41.5 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 2.9 | 17.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 2.7 | 10.7 | GO:0047298 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 2.7 | 5.4 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 2.6 | 15.9 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 2.6 | 7.8 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 2.4 | 14.4 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 2.3 | 9.0 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 2.2 | 8.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 2.1 | 8.4 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 2.1 | 21.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 2.1 | 4.2 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 2.0 | 12.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 2.0 | 6.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 2.0 | 18.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 2.0 | 44.5 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 2.0 | 6.0 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 1.9 | 19.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 1.9 | 5.6 | GO:0034584 | piRNA binding(GO:0034584) |
| 1.9 | 1.9 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 1.9 | 22.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.8 | 5.5 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 1.7 | 5.2 | GO:0070538 | oleic acid binding(GO:0070538) |
| 1.7 | 28.8 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 1.6 | 6.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 1.5 | 6.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 1.5 | 30.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 1.5 | 15.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 1.5 | 5.9 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 1.5 | 4.4 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 1.5 | 16.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.4 | 11.5 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 1.4 | 18.5 | GO:0008199 | ferric iron binding(GO:0008199) |
| 1.4 | 12.4 | GO:0043426 | MRF binding(GO:0043426) |
| 1.4 | 5.4 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 1.3 | 3.8 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 1.2 | 3.7 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 1.2 | 7.4 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 1.2 | 10.9 | GO:0045545 | syndecan binding(GO:0045545) |
| 1.2 | 29.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 1.2 | 6.0 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 1.1 | 3.4 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 1.1 | 24.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 1.1 | 8.6 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 1.0 | 10.4 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 1.0 | 6.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 1.0 | 20.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 1.0 | 3.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 1.0 | 13.3 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 1.0 | 3.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 1.0 | 12.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 1.0 | 3.0 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 1.0 | 62.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 1.0 | 8.9 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 1.0 | 38.6 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 1.0 | 7.8 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 1.0 | 9.7 | GO:0051425 | PTB domain binding(GO:0051425) |
| 1.0 | 6.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.9 | 2.8 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.9 | 4.7 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.9 | 20.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.9 | 5.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.9 | 5.4 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.9 | 91.0 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.9 | 12.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.9 | 11.3 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.9 | 2.6 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.9 | 6.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.9 | 9.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.8 | 2.5 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.8 | 12.5 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.8 | 3.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.8 | 3.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.8 | 3.2 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.8 | 3.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.8 | 3.0 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.8 | 3.0 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.7 | 2.2 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.7 | 0.7 | GO:0034618 | arginine binding(GO:0034618) |
| 0.7 | 3.5 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.7 | 4.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.7 | 5.5 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.7 | 2.0 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.7 | 3.3 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.6 | 3.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.6 | 2.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.6 | 1.8 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.6 | 59.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.6 | 17.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.6 | 9.6 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.6 | 5.3 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.6 | 1.8 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.6 | 2.3 | GO:0051870 | methotrexate binding(GO:0051870) folic acid receptor activity(GO:0061714) |
| 0.6 | 40.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.6 | 2.9 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.6 | 3.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.6 | 2.8 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.6 | 6.2 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.6 | 11.8 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.6 | 7.7 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.6 | 2.2 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.5 | 1.6 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.5 | 3.8 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.5 | 5.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.5 | 2.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.5 | 1.6 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.5 | 22.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.5 | 3.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.5 | 1.0 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.5 | 2.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.5 | 2.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.5 | 1.5 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.5 | 5.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.5 | 3.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.5 | 19.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.5 | 1.9 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.5 | 6.7 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.5 | 1.4 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.5 | 1.9 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.5 | 6.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.5 | 2.4 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.5 | 2.7 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 13.0 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.4 | 19.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.4 | 1.7 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.4 | 23.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.4 | 1.3 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.4 | 6.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.4 | 6.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.4 | 39.5 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.4 | 4.6 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.4 | 2.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.4 | 1.6 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.4 | 4.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.4 | 3.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.4 | 1.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.4 | 8.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.3 | 10.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.3 | 1.4 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.3 | 4.1 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.3 | 22.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 1.0 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.3 | 3.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 6.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 5.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.3 | 0.9 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.3 | 3.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 4.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.3 | 1.2 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.3 | 7.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.3 | 0.8 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.3 | 1.1 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.3 | 3.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.3 | 0.8 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.3 | 6.9 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.3 | 7.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.3 | 0.8 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.3 | 10.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 0.5 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.2 | 3.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 8.9 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.2 | 1.7 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 3.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 3.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.2 | 0.7 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.2 | 2.0 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 2.7 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.2 | 9.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 0.9 | GO:0016160 | amylase activity(GO:0016160) |
| 0.2 | 1.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.2 | 6.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 3.5 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.2 | 1.9 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.2 | 2.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 1.9 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.2 | 15.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 0.8 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 11.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 3.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 1.7 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.2 | 5.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.7 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.2 | 1.5 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 2.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 3.3 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.2 | 2.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 1.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 7.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 0.9 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 3.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 2.9 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 5.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.2 | 17.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 2.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 2.1 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 2.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 4.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 1.8 | GO:0019864 | IgG binding(GO:0019864) |
| 0.2 | 2.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 5.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 33.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.2 | 13.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.2 | 2.3 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.2 | 4.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.2 | 0.8 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.2 | 6.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.2 | 3.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 4.1 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 4.3 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.1 | 0.7 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.1 | 0.9 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.1 | 2.4 | GO:0016502 | G-protein coupled nucleotide receptor activity(GO:0001608) purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.0 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 1.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 4.1 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 1.0 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.5 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 3.7 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 25.9 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 3.1 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.1 | 1.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 1.4 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 4.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.5 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.1 | 7.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 10.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 2.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.4 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.1 | 2.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 3.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 1.4 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 3.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.9 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 11.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 0.5 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.1 | 0.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.6 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.9 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.6 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 0.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 1.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.2 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 1.0 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.4 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 9.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.4 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 1.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.5 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 18.0 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 8.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 3.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 2.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.8 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.1 | 0.9 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 6.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 0.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 2.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 4.3 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.1 | 18.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 0.5 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 26.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.1 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.7 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 1.3 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 1.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.8 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 1.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.6 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 3.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 4.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.7 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 1.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 2.1 | GO:0005178 | integrin binding(GO:0005178) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 19.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 1.3 | 53.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 1.2 | 71.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 1.1 | 36.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.8 | 17.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.8 | 24.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.7 | 14.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.6 | 11.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.6 | 9.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.6 | 14.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.5 | 28.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.5 | 49.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.5 | 8.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.5 | 50.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.5 | 25.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 20.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.4 | 2.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.4 | 8.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.4 | 1.9 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.4 | 93.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.3 | 12.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.3 | 5.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.3 | 2.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 13.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.3 | 8.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 7.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 2.8 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.2 | 4.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 1.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 11.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 2.0 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.2 | 7.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 8.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 6.0 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.2 | 6.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 8.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 19.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 9.2 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.2 | 8.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 1.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.2 | 3.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 8.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 3.2 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 2.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 3.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 9.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 1.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 0.7 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 1.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 1.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 0.8 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 1.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 3.9 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 2.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 21.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 0.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 1.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 1.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 2.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 3.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.5 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 3.2 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 107.0 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 2.9 | 134.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 2.5 | 5.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 1.8 | 20.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 1.2 | 23.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 1.2 | 17.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 1.0 | 23.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 1.0 | 71.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 1.0 | 15.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.9 | 22.5 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.9 | 18.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.9 | 11.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.9 | 7.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.8 | 14.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.8 | 3.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.8 | 20.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.8 | 9.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.8 | 22.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.7 | 100.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.7 | 18.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.7 | 12.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.6 | 12.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.6 | 11.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.6 | 50.8 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.6 | 29.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.6 | 33.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.5 | 20.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.5 | 7.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.5 | 11.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.4 | 9.0 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.4 | 5.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.4 | 10.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.4 | 6.6 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.4 | 7.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.4 | 2.4 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.4 | 3.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.4 | 7.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.4 | 54.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.4 | 9.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.4 | 13.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.4 | 3.3 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.4 | 28.5 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.4 | 0.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.3 | 3.4 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.3 | 10.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 7.2 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.3 | 5.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.3 | 11.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.3 | 1.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.3 | 4.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 1.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 3.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 0.7 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 4.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 5.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 3.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 1.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.2 | 42.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 3.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.2 | 2.8 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 6.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 2.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 4.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 13.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.2 | 2.1 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.2 | 1.7 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.2 | 3.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 0.6 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.1 | 8.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.3 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 2.0 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 1.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 2.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 0.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 4.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 2.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.4 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 3.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 4.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 0.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 10.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 0.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.9 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 1.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.5 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 1.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 3.0 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 2.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 4.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 1.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 4.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.9 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 2.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 8.5 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 1.1 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.5 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |