Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for STAT1_STAT3_BCL6
Z-value: 1.67
Transcription factors associated with STAT1_STAT3_BCL6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
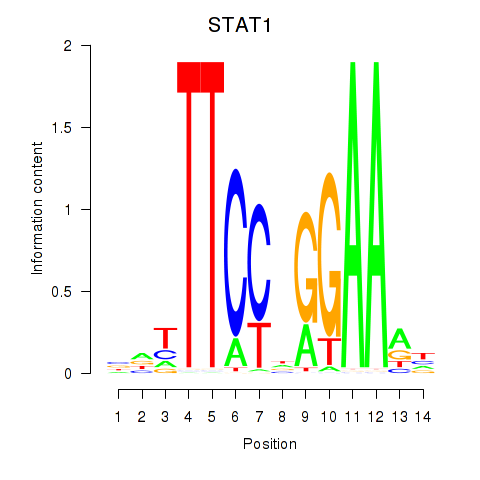
STAT1
|
ENSG00000115415.14 | signal transducer and activator of transcription 1 |
|
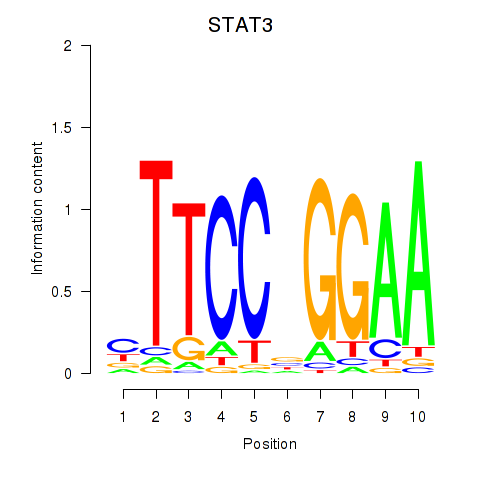
STAT3
|
ENSG00000168610.10 | signal transducer and activator of transcription 3 |
|
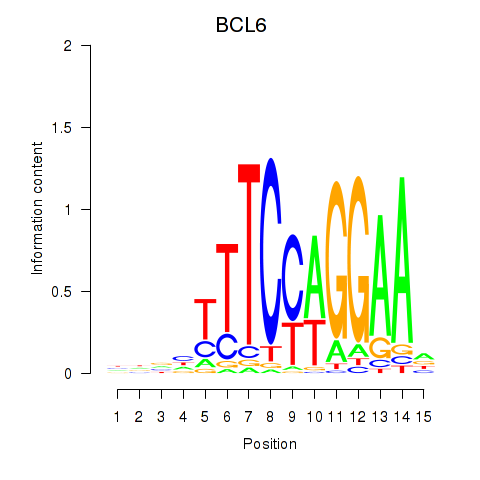
BCL6
|
ENSG00000113916.13 | BCL6 transcription repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT1 | hg19_v2_chr2_-_191878874_191878976 | -0.42 | 7.6e-11 | Click! |
| BCL6 | hg19_v2_chr3_-_187454281_187454357 | 0.29 | 1.3e-05 | Click! |
| STAT3 | hg19_v2_chr17_-_40540377_40540481 | -0.14 | 3.4e-02 | Click! |
Activity profile of STAT1_STAT3_BCL6 motif
Sorted Z-values of STAT1_STAT3_BCL6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT1_STAT3_BCL6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 23.4 | 70.3 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 21.5 | 128.8 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 13.9 | 41.8 | GO:1903410 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 12.9 | 38.8 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 8.2 | 24.6 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 6.5 | 39.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 4.2 | 16.7 | GO:0072658 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 3.6 | 10.9 | GO:0007412 | axon target recognition(GO:0007412) |
| 3.6 | 10.7 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 3.5 | 14.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 3.4 | 13.8 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 3.3 | 13.2 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 3.3 | 9.8 | GO:0048691 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 3.2 | 6.4 | GO:2001183 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 3.2 | 9.5 | GO:0072683 | T cell extravasation(GO:0072683) |
| 3.0 | 9.0 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 2.8 | 33.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 2.7 | 13.7 | GO:0030070 | insulin processing(GO:0030070) |
| 2.7 | 16.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 2.7 | 10.6 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 2.6 | 39.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 2.4 | 9.5 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 2.4 | 18.9 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 2.3 | 9.4 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 2.3 | 7.0 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 2.3 | 6.8 | GO:1903413 | cellular response to bile acid(GO:1903413) negative regulation of intestinal absorption(GO:1904479) response to iron ion starvation(GO:1990641) |
| 2.3 | 6.8 | GO:0035603 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 2.1 | 23.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 2.1 | 27.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 2.0 | 6.0 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 1.9 | 7.7 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 1.9 | 30.8 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 1.8 | 12.8 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 1.8 | 5.5 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 1.7 | 6.8 | GO:0090678 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 1.7 | 8.3 | GO:1902336 | vestibulocochlear nerve structural organization(GO:0021649) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) positive regulation of retinal ganglion cell axon guidance(GO:1902336) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 1.7 | 29.8 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 1.6 | 27.9 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 1.5 | 7.5 | GO:0044858 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 1.4 | 7.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.4 | 14.4 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 1.4 | 10.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 1.4 | 14.3 | GO:0002765 | immune response-inhibiting signal transduction(GO:0002765) immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 1.4 | 4.2 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 1.4 | 10.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 1.3 | 5.4 | GO:1905071 | ossification involved in bone remodeling(GO:0043932) frontal suture morphogenesis(GO:0060364) occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 1.3 | 4.0 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 1.3 | 6.3 | GO:0035936 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 1.2 | 12.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.2 | 4.8 | GO:0014028 | notochord formation(GO:0014028) |
| 1.1 | 3.4 | GO:0071725 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 1.1 | 3.4 | GO:1904647 | response to rotenone(GO:1904647) |
| 1.1 | 6.7 | GO:0018343 | protein farnesylation(GO:0018343) |
| 1.1 | 7.8 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 1.1 | 3.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 1.0 | 5.0 | GO:1990834 | response to odorant(GO:1990834) |
| 1.0 | 15.5 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 1.0 | 3.8 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.9 | 7.3 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.9 | 4.6 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.9 | 2.7 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.9 | 12.7 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.9 | 1.8 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.9 | 4.4 | GO:0001826 | inner cell mass cell differentiation(GO:0001826) |
| 0.9 | 8.7 | GO:0051610 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.9 | 13.0 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.8 | 2.5 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.8 | 5.9 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.8 | 10.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.8 | 17.4 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.8 | 2.4 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.8 | 4.7 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.8 | 7.0 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.8 | 3.8 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.8 | 12.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.8 | 3.8 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.8 | 8.3 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.7 | 18.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.7 | 5.6 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.7 | 4.2 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.7 | 2.7 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.7 | 4.7 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.7 | 6.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.7 | 2.7 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.7 | 2.7 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.7 | 1.3 | GO:2000570 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.7 | 4.0 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.7 | 5.9 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.7 | 3.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.6 | 6.5 | GO:0003360 | brainstem development(GO:0003360) |
| 0.6 | 7.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.6 | 5.7 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.6 | 5.7 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.6 | 0.6 | GO:0070666 | mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.6 | 38.4 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.6 | 24.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.6 | 3.5 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.6 | 4.1 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.6 | 1.1 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 0.6 | 1.7 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.6 | 3.9 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.5 | 16.3 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.5 | 10.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.5 | 2.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.5 | 8.8 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.5 | 3.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.5 | 6.6 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.5 | 2.5 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.5 | 3.5 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.5 | 1.9 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.5 | 1.9 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.5 | 1.9 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.5 | 2.9 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.5 | 7.6 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.5 | 1.9 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.5 | 8.9 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.5 | 2.8 | GO:0009624 | response to nematode(GO:0009624) |
| 0.5 | 5.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.5 | 4.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.5 | 2.3 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.4 | 2.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.4 | 6.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.4 | 1.2 | GO:0032242 | positive regulation of necrotic cell death(GO:0010940) regulation of nucleoside transport(GO:0032242) |
| 0.4 | 5.7 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.4 | 10.6 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.4 | 21.0 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.4 | 2.6 | GO:2000503 | regulation of natural killer cell chemotaxis(GO:2000501) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.4 | 1.8 | GO:0010918 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.4 | 1.1 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.4 | 5.4 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.4 | 5.3 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.4 | 2.1 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.3 | 2.8 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.3 | 13.4 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.3 | 4.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.3 | 2.0 | GO:0098700 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.3 | 1.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.3 | 1.0 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.3 | 3.0 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.3 | 5.5 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.3 | 2.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.3 | 3.3 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.3 | 9.2 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.3 | 7.7 | GO:0030220 | platelet formation(GO:0030220) |
| 0.3 | 4.1 | GO:0060315 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.3 | 3.8 | GO:0006559 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.3 | 1.4 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.3 | 2.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.3 | 4.0 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.3 | 1.6 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.3 | 2.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.3 | 1.3 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.3 | 4.9 | GO:0007567 | parturition(GO:0007567) |
| 0.3 | 4.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 2.3 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.2 | 4.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 1.7 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.2 | 3.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 2.6 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.2 | 3.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.2 | 4.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 16.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 4.8 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 0.7 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.2 | 3.6 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.2 | 2.0 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.2 | 1.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.2 | 5.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 2.4 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.2 | 3.2 | GO:0051775 | response to redox state(GO:0051775) |
| 0.2 | 1.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 0.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.2 | 1.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 10.7 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.2 | 2.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 0.8 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.2 | 5.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 3.0 | GO:0046697 | decidualization(GO:0046697) |
| 0.2 | 2.8 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.2 | 3.4 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.2 | 2.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.2 | 3.1 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.2 | 1.8 | GO:0006501 | C-terminal protein lipidation(GO:0006501) C-terminal protein amino acid modification(GO:0018410) |
| 0.2 | 1.8 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.2 | 0.9 | GO:0048852 | hypophysis morphogenesis(GO:0048850) diencephalon morphogenesis(GO:0048852) |
| 0.2 | 7.0 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.2 | 2.5 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.2 | 0.9 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.2 | 4.6 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.2 | 0.7 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.2 | 2.0 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.2 | 5.9 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.2 | 2.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.2 | 2.8 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.2 | 9.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 0.4 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.1 | 2.1 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 2.7 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 1.2 | GO:0045953 | response to yeast(GO:0001878) negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.1 | 4.3 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.1 | 1.0 | GO:0051873 | killing by host of symbiont cells(GO:0051873) |
| 0.1 | 0.3 | GO:0071503 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.1 | 0.5 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 6.0 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.5 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.1 | 1.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 4.3 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 1.8 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 11.1 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.1 | 5.8 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.1 | 5.9 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 5.7 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 5.4 | GO:0032760 | positive regulation of tumor necrosis factor production(GO:0032760) positive regulation of tumor necrosis factor superfamily cytokine production(GO:1903557) |
| 0.1 | 0.8 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 5.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 1.0 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 2.5 | GO:0035966 | response to topologically incorrect protein(GO:0035966) |
| 0.1 | 13.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 2.0 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.2 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 7.6 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 3.7 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 3.4 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 2.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 10.4 | GO:0048839 | inner ear development(GO:0048839) |
| 0.1 | 1.3 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 1.7 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 0.2 | GO:0072535 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) tumor necrosis factor (ligand) superfamily member 11 production(GO:0072535) positive regulation of bone mineralization involved in bone maturation(GO:1900159) regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000307) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 0.1 | 3.0 | GO:0006505 | GPI anchor metabolic process(GO:0006505) GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 2.5 | GO:0035904 | aorta development(GO:0035904) |
| 0.1 | 4.8 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 1.6 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 0.4 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 3.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.2 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.1 | 1.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 2.5 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 2.2 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 3.1 | GO:0030154 | cell differentiation(GO:0030154) |
| 0.1 | 0.6 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.9 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 8.0 | GO:0015992 | proton transport(GO:0015992) |
| 0.1 | 2.3 | GO:0061053 | somite development(GO:0061053) |
| 0.1 | 1.8 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 0.2 | GO:0045423 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.1 | 0.6 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.1 | 1.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 1.0 | GO:0019682 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.1 | 3.8 | GO:0007041 | lysosomal transport(GO:0007041) |
| 0.0 | 2.5 | GO:0060563 | neuroepithelial cell differentiation(GO:0060563) |
| 0.0 | 2.1 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.7 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 1.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 1.4 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 2.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.6 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 2.7 | GO:0030516 | regulation of axon extension(GO:0030516) |
| 0.0 | 0.2 | GO:1904783 | negative regulation of lamellipodium assembly(GO:0010593) regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) positive regulation of NMDA glutamate receptor activity(GO:1904783) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) regulation of barbed-end actin filament capping(GO:2000812) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 15.8 | GO:0016311 | dephosphorylation(GO:0016311) |
| 0.0 | 0.8 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 2.0 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.3 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 1.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 2.3 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.0 | 1.0 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 1.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.4 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 17.5 | GO:0043623 | cellular protein complex assembly(GO:0043623) |
| 0.0 | 4.9 | GO:0050890 | cognition(GO:0050890) |
| 0.0 | 0.6 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 2.6 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.6 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 2.4 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.4 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 | 1.3 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 6.5 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 1.7 | GO:0007088 | regulation of mitotic nuclear division(GO:0007088) |
| 0.0 | 0.3 | GO:0042692 | muscle cell differentiation(GO:0042692) |
| 0.0 | 1.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.7 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 1.4 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.1 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.8 | 39.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 8.1 | 137.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 5.0 | 25.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 3.7 | 11.1 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 3.5 | 53.0 | GO:0030478 | actin cap(GO:0030478) |
| 3.1 | 9.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 3.0 | 15.0 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 2.6 | 29.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 2.4 | 33.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 2.3 | 39.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 2.1 | 10.7 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 2.1 | 14.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 2.0 | 10.1 | GO:0001652 | granular component(GO:0001652) |
| 1.8 | 29.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.8 | 7.0 | GO:0031673 | H zone(GO:0031673) |
| 1.7 | 6.7 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 1.7 | 14.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.5 | 6.1 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 1.5 | 4.4 | GO:0016938 | kinesin I complex(GO:0016938) |
| 1.5 | 23.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 1.3 | 16.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.2 | 7.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 1.1 | 18.9 | GO:0032059 | bleb(GO:0032059) |
| 1.0 | 8.4 | GO:0045179 | apical cortex(GO:0045179) |
| 1.0 | 6.8 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.9 | 29.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.9 | 4.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.8 | 5.7 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.8 | 8.8 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.7 | 6.7 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.7 | 4.2 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.6 | 5.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.6 | 1.8 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.6 | 10.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.5 | 3.8 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.5 | 7.8 | GO:0042627 | chylomicron(GO:0042627) |
| 0.5 | 8.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.5 | 5.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.5 | 53.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.5 | 5.9 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.4 | 13.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.4 | 14.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.4 | 10.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.3 | 2.0 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.3 | 12.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.3 | 5.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 3.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.3 | 17.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.3 | 4.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 12.6 | GO:0031672 | A band(GO:0031672) |
| 0.3 | 46.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 3.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.2 | 15.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 3.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 3.2 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.2 | 114.1 | GO:0005813 | centrosome(GO:0005813) |
| 0.2 | 13.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 2.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 1.9 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 2.0 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.2 | 2.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 10.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 3.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 7.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 3.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 3.5 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 14.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 8.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 4.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 7.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 30.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 3.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 7.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 5.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 2.5 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 1.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 5.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 1.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 13.8 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.1 | 1.0 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 7.4 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 2.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 6.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 0.2 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 5.0 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.1 | 14.2 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 0.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 5.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 1.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 0.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 1.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.7 | GO:0042589 | zymogen granule(GO:0042588) zymogen granule membrane(GO:0042589) |
| 0.1 | 4.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 9.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 11.4 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 2.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 7.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 2.8 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 2.8 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 13.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 50.3 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 3.1 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.6 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.1 | GO:0031012 | extracellular matrix(GO:0031012) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.1 | 48.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 13.8 | 41.3 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 8.9 | 35.6 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 8.8 | 70.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 6.4 | 128.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 5.6 | 33.6 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 2.7 | 10.9 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 2.6 | 7.8 | GO:0017129 | triglyceride binding(GO:0017129) |
| 2.5 | 24.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 2.5 | 41.8 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 2.3 | 9.4 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 2.2 | 13.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 2.2 | 10.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.9 | 5.7 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 1.8 | 5.5 | GO:0070538 | oleic acid binding(GO:0070538) |
| 1.8 | 23.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 1.8 | 5.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 1.8 | 10.7 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.7 | 6.7 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 1.6 | 19.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 1.5 | 9.0 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 1.5 | 6.0 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 1.5 | 41.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.5 | 8.8 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 1.4 | 14.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 1.4 | 8.6 | GO:0030109 | HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) |
| 1.4 | 16.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.2 | 16.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.2 | 19.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 1.2 | 15.9 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 1.2 | 7.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 1.1 | 5.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.9 | 6.6 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.9 | 3.8 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.9 | 29.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.9 | 8.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.9 | 5.4 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.9 | 10.8 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.9 | 11.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.8 | 2.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.8 | 5.7 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.8 | 10.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) neurotrophin receptor activity(GO:0005030) |
| 0.8 | 3.8 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.8 | 8.3 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.8 | 2.3 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.8 | 6.0 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.7 | 10.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.7 | 2.2 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.7 | 5.1 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.7 | 7.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.7 | 2.9 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.7 | 2.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.7 | 19.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.7 | 2.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.7 | 2.0 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.7 | 6.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.7 | 6.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.6 | 7.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.6 | 2.5 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.6 | 6.8 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.6 | 4.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.6 | 3.5 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.6 | 10.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.6 | 9.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.6 | 1.7 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.6 | 2.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.5 | 4.9 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.5 | 2.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.5 | 1.6 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.5 | 13.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.5 | 2.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.5 | 2.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.5 | 45.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.5 | 4.4 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.5 | 2.4 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.5 | 2.8 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.4 | 4.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.4 | 3.5 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.4 | 3.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.4 | 2.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.4 | 5.7 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.4 | 4.0 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.4 | 10.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.4 | 3.5 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.4 | 18.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.4 | 2.2 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.4 | 4.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.4 | 4.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.3 | 3.5 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.3 | 1.3 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.3 | 1.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.3 | 7.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.3 | 6.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.3 | 3.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 4.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.3 | 30.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 3.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.3 | 6.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 6.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.3 | 2.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.3 | 1.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.3 | 2.5 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.3 | 2.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 4.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 3.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 7.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 5.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 2.7 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.2 | 2.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 4.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.2 | 1.7 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 1.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 31.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 1.0 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 6.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 1.6 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 1.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 1.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 7.0 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.2 | 2.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 5.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 7.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 3.8 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.2 | 2.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 14.7 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.2 | 1.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 2.6 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.2 | 0.9 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.2 | 5.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 2.6 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 1.7 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.2 | 7.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 1.7 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.1 | 3.4 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 16.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.8 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 4.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 3.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.0 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.7 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.1 | 0.5 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 4.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 10.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 2.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 2.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 8.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 10.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 3.3 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 1.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 9.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 3.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 3.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 1.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 1.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 4.6 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 12.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 3.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 3.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.6 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.9 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 1.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 6.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 16.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 10.6 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 42.8 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.1 | 0.5 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.3 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 10.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.1 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 14.8 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 2.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 19.5 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 1.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 6.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 3.2 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 41.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 1.1 | 75.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.8 | 20.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.8 | 6.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.8 | 54.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.8 | 41.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.8 | 101.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.7 | 10.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.6 | 39.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.6 | 8.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.5 | 7.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.4 | 7.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.4 | 5.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.4 | 1.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.4 | 15.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.4 | 63.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.4 | 13.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.4 | 13.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.3 | 74.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.3 | 14.7 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.3 | 7.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.3 | 8.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.3 | 7.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 9.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 5.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.2 | 11.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 9.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.2 | 15.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.2 | 10.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.2 | 5.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 1.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 5.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.2 | 4.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 6.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 5.9 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.2 | 1.5 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.2 | 4.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 10.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 13.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 8.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 2.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.0 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 2.3 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 0.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 30.1 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.1 | 1.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 7.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.8 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 1.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 2.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.3 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 43.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 2.2 | 24.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.9 | 23.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 1.7 | 107.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 1.2 | 33.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 1.2 | 21.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 1.1 | 35.0 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 1.1 | 142.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 1.0 | 34.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.8 | 19.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.8 | 17.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.8 | 8.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.6 | 35.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.6 | 3.5 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.5 | 9.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.4 | 53.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.4 | 5.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.4 | 6.8 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.4 | 13.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.4 | 3.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.4 | 14.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.4 | 9.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.4 | 7.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 2.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 11.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.3 | 62.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.3 | 4.0 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 2.9 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.3 | 6.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.3 | 1.8 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.3 | 6.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.2 | 9.7 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.2 | 6.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 2.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 3.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 6.5 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.2 | 4.9 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 3.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 3.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 2.5 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 3.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 4.2 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.2 | 9.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.2 | 9.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 7.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 3.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 5.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 3.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 4.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 2.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 2.9 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 13.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 8.9 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.1 | 2.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 2.5 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 8.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 4.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 2.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 3.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 3.0 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.1 | 2.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 2.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 3.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 2.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.0 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.1 | 0.6 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 1.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.6 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.1 | 1.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 2.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 2.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 3.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 1.8 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |