Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
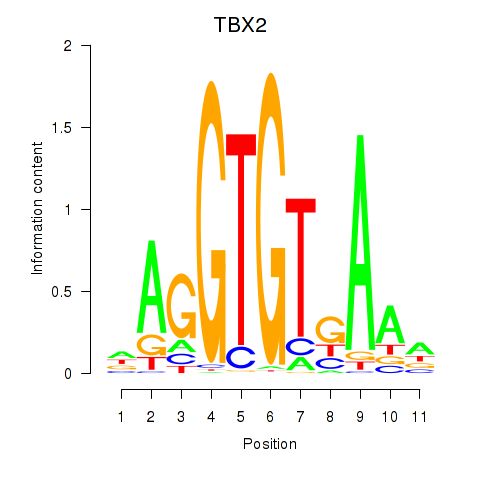
Results for TBX2
Z-value: 0.79
Transcription factors associated with TBX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX2
|
ENSG00000121068.9 | T-box transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX2 | hg19_v2_chr17_+_59477233_59477263 | -0.18 | 6.5e-03 | Click! |
Activity profile of TBX2 motif
Sorted Z-values of TBX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_113247543 | 13.54 |
ENST00000414971.1
ENST00000534717.1 |
RHOC
|
ras homolog family member C |
| chr8_+_82192501 | 13.12 |
ENST00000297258.6
|
FABP5
|
fatty acid binding protein 5 (psoriasis-associated) |
| chr17_+_35849937 | 12.80 |
ENST00000394389.4
|
DUSP14
|
dual specificity phosphatase 14 |
| chr7_+_141438393 | 11.52 |
ENST00000484178.1
ENST00000473783.1 ENST00000481508.1 |
SSBP1
|
single-stranded DNA binding protein 1, mitochondrial |
| chrX_+_70503037 | 11.44 |
ENST00000535149.1
|
NONO
|
non-POU domain containing, octamer-binding |
| chr2_+_201994042 | 11.35 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr19_+_50180317 | 11.10 |
ENST00000534465.1
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr12_-_56121580 | 11.07 |
ENST00000550776.1
|
CD63
|
CD63 molecule |
| chr9_+_131447342 | 10.87 |
ENST00000409104.3
|
SET
|
SET nuclear oncogene |
| chrX_-_53461288 | 10.87 |
ENST00000375298.4
ENST00000375304.5 |
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr16_-_66968055 | 10.68 |
ENST00000568572.1
|
FAM96B
|
family with sequence similarity 96, member B |
| chr12_+_96252706 | 10.54 |
ENST00000266735.5
ENST00000553192.1 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr7_+_73106926 | 10.51 |
ENST00000453316.1
|
WBSCR22
|
Williams Beuren syndrome chromosome region 22 |
| chr10_+_54074033 | 9.94 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr9_+_131452239 | 9.88 |
ENST00000372688.4
ENST00000372686.5 |
SET
|
SET nuclear oncogene |
| chr12_-_56121612 | 9.19 |
ENST00000546939.1
|
CD63
|
CD63 molecule |
| chrX_-_53461305 | 8.96 |
ENST00000168216.6
|
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr19_+_36236491 | 8.94 |
ENST00000591949.1
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr12_-_56120838 | 8.69 |
ENST00000548160.1
|
CD63
|
CD63 molecule |
| chr19_+_52932435 | 8.66 |
ENST00000301085.4
|
ZNF534
|
zinc finger protein 534 |
| chr11_-_14380664 | 8.51 |
ENST00000545643.1
ENST00000256196.4 |
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr19_-_51875894 | 8.47 |
ENST00000600427.1
ENST00000595217.1 ENST00000221978.5 |
NKG7
|
natural killer cell group 7 sequence |
| chr12_-_56122220 | 8.28 |
ENST00000552692.1
|
CD63
|
CD63 molecule |
| chr16_-_88717423 | 8.09 |
ENST00000568278.1
ENST00000569359.1 ENST00000567174.1 |
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr3_+_160117087 | 8.07 |
ENST00000357388.3
|
SMC4
|
structural maintenance of chromosomes 4 |
| chr19_+_47634039 | 7.99 |
ENST00000597808.1
ENST00000413379.3 ENST00000600706.1 ENST00000540850.1 ENST00000598840.1 ENST00000600753.1 ENST00000270225.7 ENST00000392776.3 |
SAE1
|
SUMO1 activating enzyme subunit 1 |
| chr15_-_55581954 | 7.96 |
ENST00000336787.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr19_+_36236514 | 7.96 |
ENST00000222266.2
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr1_+_15736359 | 7.73 |
ENST00000375980.4
|
EFHD2
|
EF-hand domain family, member D2 |
| chr14_-_55658252 | 7.45 |
ENST00000395425.2
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr14_-_100841670 | 7.40 |
ENST00000557297.1
ENST00000555813.1 ENST00000557135.1 ENST00000556698.1 ENST00000554509.1 ENST00000555410.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr10_+_95256356 | 7.35 |
ENST00000371485.3
|
CEP55
|
centrosomal protein 55kDa |
| chr13_-_46543805 | 7.34 |
ENST00000378921.2
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr19_-_47353547 | 7.15 |
ENST00000601498.1
|
AP2S1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr5_+_52856456 | 7.13 |
ENST00000296684.5
ENST00000506765.1 |
NDUFS4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase) |
| chr2_-_169769787 | 7.11 |
ENST00000451987.1
|
SPC25
|
SPC25, NDC80 kinetochore complex component |
| chr11_-_6704513 | 7.07 |
ENST00000532203.1
ENST00000288937.6 |
MRPL17
|
mitochondrial ribosomal protein L17 |
| chr1_+_93544821 | 6.95 |
ENST00000370303.4
|
MTF2
|
metal response element binding transcription factor 2 |
| chr2_+_201994208 | 6.91 |
ENST00000440180.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr14_-_55658323 | 6.89 |
ENST00000554067.1
ENST00000247191.2 |
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr12_-_56122124 | 6.75 |
ENST00000552754.1
|
CD63
|
CD63 molecule |
| chr3_+_142315225 | 6.73 |
ENST00000457734.2
ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1
|
plastin 1 |
| chr12_-_49318715 | 6.63 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr7_-_76255444 | 6.52 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr16_-_67493110 | 6.46 |
ENST00000602876.1
|
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr12_-_56122426 | 6.42 |
ENST00000551173.1
|
CD63
|
CD63 molecule |
| chr4_+_140222609 | 6.33 |
ENST00000296543.5
ENST00000398947.1 |
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr12_-_121019165 | 6.31 |
ENST00000341039.2
ENST00000357500.4 |
POP5
|
processing of precursor 5, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr14_-_24664540 | 6.30 |
ENST00000530563.1
ENST00000528895.1 ENST00000528669.1 ENST00000532632.1 |
TM9SF1
|
transmembrane 9 superfamily member 1 |
| chr19_+_50180409 | 6.30 |
ENST00000391851.4
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr14_-_45603657 | 6.27 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr5_+_154320623 | 6.26 |
ENST00000523037.1
ENST00000265229.8 ENST00000439747.3 ENST00000522038.1 |
MRPL22
|
mitochondrial ribosomal protein L22 |
| chr1_+_32716840 | 6.26 |
ENST00000336890.5
|
LCK
|
lymphocyte-specific protein tyrosine kinase |
| chr20_+_25388293 | 6.21 |
ENST00000262460.4
ENST00000429262.2 |
GINS1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr6_-_160166218 | 6.17 |
ENST00000537657.1
|
SOD2
|
superoxide dismutase 2, mitochondrial |
| chr8_-_67974552 | 6.06 |
ENST00000357849.4
|
COPS5
|
COP9 signalosome subunit 5 |
| chr1_-_6260896 | 6.02 |
ENST00000497965.1
|
RPL22
|
ribosomal protein L22 |
| chr11_+_108535752 | 6.00 |
ENST00000322536.3
|
DDX10
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
| chrX_+_114795489 | 6.00 |
ENST00000355899.3
ENST00000537301.1 ENST00000289290.3 |
PLS3
|
plastin 3 |
| chr7_+_100271446 | 5.99 |
ENST00000419828.1
ENST00000427895.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr19_+_50180507 | 5.96 |
ENST00000454376.2
ENST00000524771.1 |
PRMT1
|
protein arginine methyltransferase 1 |
| chr12_-_111180644 | 5.93 |
ENST00000551676.1
ENST00000550991.1 ENST00000335007.5 ENST00000340766.5 |
PPP1CC
|
protein phosphatase 1, catalytic subunit, gamma isozyme |
| chr1_+_32716857 | 5.86 |
ENST00000482949.1
ENST00000495610.2 |
LCK
|
lymphocyte-specific protein tyrosine kinase |
| chr3_+_112709804 | 5.85 |
ENST00000383677.3
|
GTPBP8
|
GTP-binding protein 8 (putative) |
| chr17_-_7590745 | 5.79 |
ENST00000514944.1
ENST00000503591.1 ENST00000455263.2 ENST00000420246.2 ENST00000445888.2 ENST00000509690.1 ENST00000604348.1 ENST00000269305.4 |
TP53
|
tumor protein p53 |
| chr17_+_48823896 | 5.70 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr19_+_13049413 | 5.61 |
ENST00000316448.5
ENST00000588454.1 |
CALR
|
calreticulin |
| chr9_-_179018 | 5.59 |
ENST00000431099.2
ENST00000382447.4 ENST00000382389.1 ENST00000377447.3 ENST00000314367.10 ENST00000356521.4 ENST00000382393.1 ENST00000377400.4 |
CBWD1
|
COBW domain containing 1 |
| chr11_-_6633799 | 5.58 |
ENST00000299424.4
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa |
| chr7_+_872107 | 5.49 |
ENST00000405266.1
ENST00000401592.1 ENST00000403868.1 ENST00000425407.2 |
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr12_-_56122761 | 5.45 |
ENST00000552164.1
ENST00000420846.3 ENST00000257857.4 |
CD63
|
CD63 molecule |
| chr2_-_209118974 | 5.40 |
ENST00000415913.1
ENST00000415282.1 ENST00000446179.1 |
IDH1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr9_-_127177703 | 5.39 |
ENST00000259457.3
ENST00000536392.1 ENST00000441097.1 |
PSMB7
|
proteasome (prosome, macropain) subunit, beta type, 7 |
| chr2_+_136343820 | 5.37 |
ENST00000410054.1
|
R3HDM1
|
R3H domain containing 1 |
| chr19_-_51875523 | 5.37 |
ENST00000593572.1
ENST00000595157.1 |
NKG7
|
natural killer cell group 7 sequence |
| chr11_+_114270752 | 5.35 |
ENST00000540163.1
|
RBM7
|
RNA binding motif protein 7 |
| chr1_+_156105878 | 5.20 |
ENST00000508500.1
|
LMNA
|
lamin A/C |
| chr2_+_219081817 | 5.15 |
ENST00000315717.5
ENST00000420104.1 ENST00000295685.10 |
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr16_-_88717482 | 5.12 |
ENST00000261623.3
|
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr14_-_51411194 | 5.07 |
ENST00000544180.2
|
PYGL
|
phosphorylase, glycogen, liver |
| chr7_+_107220899 | 5.05 |
ENST00000379117.2
ENST00000473124.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr21_+_30502806 | 4.91 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr17_-_39684550 | 4.85 |
ENST00000455635.1
ENST00000361566.3 |
KRT19
|
keratin 19 |
| chr8_-_124428569 | 4.82 |
ENST00000521903.1
|
ATAD2
|
ATPase family, AAA domain containing 2 |
| chr13_+_34392185 | 4.73 |
ENST00000380071.3
|
RFC3
|
replication factor C (activator 1) 3, 38kDa |
| chr14_+_90864504 | 4.73 |
ENST00000544280.2
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr1_-_146040968 | 4.70 |
ENST00000401010.3
|
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chrX_-_10645773 | 4.69 |
ENST00000453318.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr12_-_56120865 | 4.66 |
ENST00000548898.1
ENST00000552067.1 |
CD63
|
CD63 molecule |
| chr5_-_9630463 | 4.61 |
ENST00000382492.2
|
TAS2R1
|
taste receptor, type 2, member 1 |
| chr11_+_114271251 | 4.61 |
ENST00000375490.5
|
RBM7
|
RNA binding motif protein 7 |
| chr11_-_58345569 | 4.54 |
ENST00000528954.1
ENST00000528489.1 |
LPXN
|
leupaxin |
| chr1_-_205744205 | 4.52 |
ENST00000446390.2
|
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr6_+_32936353 | 4.51 |
ENST00000374825.4
|
BRD2
|
bromodomain containing 2 |
| chr7_+_107220660 | 4.50 |
ENST00000465919.1
ENST00000445771.2 ENST00000479917.1 ENST00000421217.1 ENST00000457837.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr8_+_1993173 | 4.49 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr12_+_21525818 | 4.49 |
ENST00000240652.3
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chr11_-_615570 | 4.44 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr19_-_14628645 | 4.35 |
ENST00000598235.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr14_-_75643296 | 4.34 |
ENST00000303575.4
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast) |
| chr12_+_75874984 | 4.28 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr10_+_60028818 | 4.22 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr20_-_24973318 | 4.14 |
ENST00000447138.1
|
APMAP
|
adipocyte plasma membrane associated protein |
| chr11_+_69455855 | 4.13 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr5_-_71616043 | 4.12 |
ENST00000508863.2
ENST00000522095.1 ENST00000513900.1 ENST00000515404.1 ENST00000457646.4 ENST00000261413.5 |
MRPS27
|
mitochondrial ribosomal protein S27 |
| chr8_-_49834299 | 4.12 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr16_-_4395905 | 4.12 |
ENST00000571941.1
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr19_-_41942344 | 4.08 |
ENST00000594660.1
|
ATP5SL
|
ATP5S-like |
| chrX_-_70329118 | 4.02 |
ENST00000374188.3
|
IL2RG
|
interleukin 2 receptor, gamma |
| chr9_-_70488865 | 4.01 |
ENST00000377392.5
|
CBWD5
|
COBW domain containing 5 |
| chr5_+_82767583 | 3.91 |
ENST00000512590.2
ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN
|
versican |
| chrX_+_51636629 | 3.91 |
ENST00000375722.1
ENST00000326587.7 ENST00000375695.2 |
MAGED1
|
melanoma antigen family D, 1 |
| chr3_+_112709755 | 3.78 |
ENST00000383678.2
|
GTPBP8
|
GTP-binding protein 8 (putative) |
| chr2_-_85581701 | 3.73 |
ENST00000295802.4
|
RETSAT
|
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr11_-_414948 | 3.73 |
ENST00000530494.1
ENST00000528209.1 ENST00000431843.2 ENST00000528058.1 |
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr7_+_148892557 | 3.72 |
ENST00000262085.3
|
ZNF282
|
zinc finger protein 282 |
| chr13_+_114238997 | 3.70 |
ENST00000538138.1
ENST00000375370.5 |
TFDP1
|
transcription factor Dp-1 |
| chr19_-_45982076 | 3.65 |
ENST00000423698.2
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr16_-_30440530 | 3.62 |
ENST00000568434.1
|
DCTPP1
|
dCTP pyrophosphatase 1 |
| chr12_+_19358228 | 3.56 |
ENST00000424268.1
ENST00000543806.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr1_-_25256368 | 3.51 |
ENST00000308873.6
|
RUNX3
|
runt-related transcription factor 3 |
| chr3_+_184033135 | 3.51 |
ENST00000424196.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr3_-_183967296 | 3.46 |
ENST00000455059.1
ENST00000445626.2 |
ALG3
|
ALG3, alpha-1,3- mannosyltransferase |
| chr22_-_31328881 | 3.37 |
ENST00000445980.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chrX_-_7066159 | 3.36 |
ENST00000486446.2
ENST00000412827.2 ENST00000424830.2 ENST00000381077.5 ENST00000540122.1 |
HDHD1
|
haloacid dehalogenase-like hydrolase domain containing 1 |
| chr2_+_169312725 | 3.35 |
ENST00000392687.4
|
CERS6
|
ceramide synthase 6 |
| chr12_+_1800179 | 3.33 |
ENST00000357103.4
|
ADIPOR2
|
adiponectin receptor 2 |
| chr6_+_30585486 | 3.32 |
ENST00000259873.4
ENST00000506373.2 |
MRPS18B
|
mitochondrial ribosomal protein S18B |
| chr12_-_49351228 | 3.31 |
ENST00000541959.1
ENST00000447318.2 |
ARF3
|
ADP-ribosylation factor 3 |
| chr14_+_103058948 | 3.21 |
ENST00000262241.6
|
RCOR1
|
REST corepressor 1 |
| chr15_-_52483566 | 3.21 |
ENST00000261837.7
|
GNB5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr19_+_55897699 | 3.18 |
ENST00000558131.1
ENST00000558752.1 ENST00000458349.2 |
RPL28
|
ribosomal protein L28 |
| chr2_-_191878874 | 3.18 |
ENST00000392322.3
ENST00000392323.2 ENST00000424722.1 ENST00000361099.3 |
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr3_-_79068594 | 3.13 |
ENST00000436010.2
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr1_-_211752073 | 3.12 |
ENST00000367001.4
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1 |
| chr11_-_28129656 | 3.12 |
ENST00000263181.6
|
KIF18A
|
kinesin family member 18A |
| chr8_+_21823726 | 3.06 |
ENST00000433566.4
|
XPO7
|
exportin 7 |
| chr1_+_20915409 | 3.05 |
ENST00000375071.3
|
CDA
|
cytidine deaminase |
| chr3_-_120170052 | 3.05 |
ENST00000295633.3
|
FSTL1
|
follistatin-like 1 |
| chr11_-_66112555 | 3.04 |
ENST00000425825.2
ENST00000359957.3 |
BRMS1
|
breast cancer metastasis suppressor 1 |
| chr9_+_35829208 | 3.03 |
ENST00000439587.2
ENST00000377991.4 |
TMEM8B
|
transmembrane protein 8B |
| chr5_+_35856951 | 3.00 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr19_-_44174330 | 2.96 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr2_+_234621551 | 2.96 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr16_+_30212378 | 2.95 |
ENST00000569485.1
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr22_+_24823517 | 2.91 |
ENST00000496258.1
ENST00000337539.7 |
ADORA2A
|
adenosine A2a receptor |
| chr1_+_93544791 | 2.89 |
ENST00000545708.1
ENST00000540243.1 ENST00000370298.4 |
MTF2
|
metal response element binding transcription factor 2 |
| chr19_-_39108552 | 2.87 |
ENST00000591517.1
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr19_-_43702231 | 2.86 |
ENST00000597374.1
ENST00000599371.1 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr12_-_49351303 | 2.85 |
ENST00000256682.4
|
ARF3
|
ADP-ribosylation factor 3 |
| chrX_-_119709637 | 2.85 |
ENST00000404115.3
|
CUL4B
|
cullin 4B |
| chr12_+_112279782 | 2.82 |
ENST00000550735.2
|
MAPKAPK5
|
mitogen-activated protein kinase-activated protein kinase 5 |
| chr5_-_137514333 | 2.78 |
ENST00000411594.2
ENST00000430331.1 |
BRD8
|
bromodomain containing 8 |
| chr14_-_104028595 | 2.77 |
ENST00000337322.4
ENST00000445922.2 |
BAG5
|
BCL2-associated athanogene 5 |
| chr20_-_21378666 | 2.71 |
ENST00000351817.4
|
NKX2-4
|
NK2 homeobox 4 |
| chr3_+_157827841 | 2.71 |
ENST00000295930.3
ENST00000471994.1 ENST00000464171.1 ENST00000312179.6 ENST00000475278.2 |
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr8_-_99954788 | 2.69 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr6_+_30035307 | 2.65 |
ENST00000376765.2
ENST00000376763.1 |
PPP1R11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr1_+_8021713 | 2.63 |
ENST00000338639.5
ENST00000493678.1 ENST00000377493.5 |
PARK7
|
parkinson protein 7 |
| chr22_+_40441456 | 2.62 |
ENST00000402203.1
|
TNRC6B
|
trinucleotide repeat containing 6B |
| chr3_-_125094093 | 2.61 |
ENST00000484491.1
ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148
|
zinc finger protein 148 |
| chr22_+_23522552 | 2.58 |
ENST00000359540.3
ENST00000398512.5 |
BCR
|
breakpoint cluster region |
| chr2_+_189156586 | 2.57 |
ENST00000409830.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr11_+_237016 | 2.55 |
ENST00000352303.5
|
PSMD13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr11_+_122709200 | 2.54 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr19_-_39108643 | 2.53 |
ENST00000396857.2
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr2_+_26467825 | 2.52 |
ENST00000545822.1
ENST00000425035.1 |
HADHB
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit |
| chr1_-_200589859 | 2.51 |
ENST00000367350.4
|
KIF14
|
kinesin family member 14 |
| chr13_-_48669232 | 2.48 |
ENST00000258648.2
ENST00000378586.1 |
MED4
|
mediator complex subunit 4 |
| chr7_-_87849340 | 2.48 |
ENST00000419179.1
ENST00000265729.2 |
SRI
|
sorcin |
| chr10_-_5060147 | 2.47 |
ENST00000604507.1
|
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chr17_+_28804380 | 2.47 |
ENST00000225724.5
ENST00000451249.2 ENST00000467337.2 ENST00000581721.1 ENST00000414833.2 |
GOSR1
|
golgi SNAP receptor complex member 1 |
| chr11_-_85779971 | 2.46 |
ENST00000393346.3
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr1_+_161736072 | 2.45 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr10_-_112678904 | 2.45 |
ENST00000423273.1
ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1
|
BBSome interacting protein 1 |
| chr7_-_35077653 | 2.42 |
ENST00000310974.4
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr17_-_61996136 | 2.35 |
ENST00000342364.4
|
GH1
|
growth hormone 1 |
| chr3_+_42190714 | 2.35 |
ENST00000449246.1
|
TRAK1
|
trafficking protein, kinesin binding 1 |
| chr2_+_26467762 | 2.33 |
ENST00000317799.5
ENST00000405867.3 ENST00000537713.1 |
HADHB
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit |
| chr3_-_119379719 | 2.31 |
ENST00000493094.1
|
POPDC2
|
popeye domain containing 2 |
| chr12_-_57522813 | 2.31 |
ENST00000556155.1
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr11_+_236959 | 2.29 |
ENST00000431206.2
ENST00000528906.1 |
PSMD13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr1_+_44444865 | 2.28 |
ENST00000372324.1
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chrX_+_123480194 | 2.25 |
ENST00000371139.4
|
SH2D1A
|
SH2 domain containing 1A |
| chr10_-_112678692 | 2.24 |
ENST00000605742.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr11_+_20409070 | 2.12 |
ENST00000331079.6
|
PRMT3
|
protein arginine methyltransferase 3 |
| chr2_-_136743169 | 2.11 |
ENST00000264161.4
|
DARS
|
aspartyl-tRNA synthetase |
| chr22_+_38597889 | 2.11 |
ENST00000338483.2
ENST00000538320.1 ENST00000538999.1 ENST00000441709.1 |
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr5_-_176433350 | 2.11 |
ENST00000377227.4
ENST00000377219.2 |
UIMC1
|
ubiquitin interaction motif containing 1 |
| chr17_+_34538310 | 2.11 |
ENST00000444414.1
ENST00000378350.4 ENST00000389068.5 ENST00000588929.1 ENST00000589079.1 ENST00000589336.1 ENST00000400702.4 ENST00000591167.1 ENST00000586598.1 ENST00000591637.1 ENST00000378352.4 ENST00000358756.5 |
CCL4L1
|
chemokine (C-C motif) ligand 4-like 1 |
| chr1_-_205290865 | 2.10 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chr2_-_26467557 | 2.10 |
ENST00000380649.3
|
HADHA
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit |
| chr16_+_67906919 | 2.09 |
ENST00000358933.5
|
EDC4
|
enhancer of mRNA decapping 4 |
| chr2_-_40679186 | 2.01 |
ENST00000406785.2
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr16_+_12059091 | 2.01 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr22_-_41215291 | 2.00 |
ENST00000542412.1
ENST00000544408.1 |
SLC25A17
|
solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 |
| chr11_-_61735103 | 1.98 |
ENST00000529191.1
ENST00000529631.1 ENST00000530019.1 ENST00000529548.1 ENST00000273550.7 |
FTH1
|
ferritin, heavy polypeptide 1 |
| chr3_-_105588231 | 1.97 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr6_+_43603552 | 1.97 |
ENST00000372171.4
|
MAD2L1BP
|
MAD2L1 binding protein |
| chr14_-_23904861 | 1.96 |
ENST00000355349.3
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.6 | 19.8 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 4.7 | 60.5 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 3.9 | 23.4 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 3.3 | 9.9 | GO:0009996 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) negative regulation of cell fate specification(GO:0009996) Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 2.6 | 13.2 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 2.4 | 14.3 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 2.3 | 14.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 2.3 | 13.5 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 1.9 | 5.8 | GO:0051097 | negative regulation of helicase activity(GO:0051097) oligodendrocyte apoptotic process(GO:0097252) |
| 1.8 | 18.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 1.8 | 5.4 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 1.6 | 9.8 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 1.5 | 7.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 1.4 | 4.1 | GO:1901420 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) negative regulation of response to alcohol(GO:1901420) |
| 1.4 | 5.5 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 1.3 | 6.7 | GO:1902896 | terminal web assembly(GO:1902896) |
| 1.3 | 8.0 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 1.2 | 6.2 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 1.2 | 6.1 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 1.2 | 6.9 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 1.2 | 11.5 | GO:0051095 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 1.1 | 6.5 | GO:0035803 | egg coat formation(GO:0035803) |
| 1.1 | 4.3 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 1.1 | 3.2 | GO:0061317 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) arterial endothelial cell fate commitment(GO:0060844) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) blood vessel lumenization(GO:0072554) positive regulation of ephrin receptor signaling pathway(GO:1901189) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 1.1 | 9.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 1.1 | 6.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 1.1 | 4.2 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 1.0 | 3.1 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 1.0 | 5.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 1.0 | 4.0 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 1.0 | 8.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 1.0 | 2.9 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 1.0 | 18.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.9 | 4.7 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.9 | 5.6 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.9 | 2.6 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.8 | 2.4 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.8 | 3.2 | GO:2000697 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.8 | 12.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.8 | 4.5 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.7 | 3.7 | GO:0000720 | meiotic mismatch repair(GO:0000710) pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.7 | 3.6 | GO:0009213 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.7 | 5.8 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.7 | 18.8 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.7 | 2.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.6 | 4.5 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.6 | 4.4 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.6 | 6.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.6 | 1.9 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.6 | 3.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.6 | 6.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.6 | 1.8 | GO:0006788 | heme oxidation(GO:0006788) smooth muscle hyperplasia(GO:0014806) |
| 0.6 | 3.0 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.6 | 4.0 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.6 | 1.7 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.6 | 1.7 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.5 | 4.8 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.5 | 3.7 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.5 | 4.8 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.5 | 2.5 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.5 | 2.0 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.5 | 2.5 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.5 | 10.7 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.5 | 2.8 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.4 | 3.5 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.4 | 5.6 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.4 | 6.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.4 | 1.3 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.4 | 2.9 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.4 | 3.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.4 | 4.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.4 | 7.4 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.4 | 6.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.4 | 6.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.4 | 7.2 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.4 | 3.0 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.4 | 2.5 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.4 | 4.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.4 | 4.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.4 | 2.5 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.4 | 2.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.3 | 1.7 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.3 | 2.7 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.3 | 3.6 | GO:0061458 | reproductive system development(GO:0061458) |
| 0.3 | 11.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.3 | 1.6 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.3 | 2.8 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.3 | 6.5 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.3 | 2.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.3 | 6.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 13.5 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.3 | 3.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.3 | 13.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.3 | 2.9 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.3 | 20.8 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.3 | 0.6 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.3 | 0.8 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.3 | 4.1 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.3 | 2.9 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.3 | 2.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.3 | 2.4 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.3 | 2.0 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.2 | 0.5 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.2 | 3.1 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.2 | 5.2 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.2 | 1.4 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.2 | 3.9 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 | 10.3 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.2 | 2.0 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.2 | 2.3 | GO:0002829 | negative regulation of type 2 immune response(GO:0002829) |
| 0.2 | 2.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.2 | 3.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 1.6 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 0.6 | GO:0043132 | FAD transport(GO:0015883) FAD transmembrane transport(GO:0035350) NAD transport(GO:0043132) |
| 0.2 | 2.0 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.2 | 4.8 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.2 | 0.6 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.2 | 0.5 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.2 | 5.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 2.6 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.2 | 0.7 | GO:1904379 | maintenance of unfolded protein(GO:0036506) protein localization to cytosolic proteasome complex(GO:1904327) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.2 | 17.4 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.2 | 2.4 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.2 | 4.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 12.1 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.2 | 0.8 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 4.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 6.3 | GO:0006914 | autophagy(GO:0006914) |
| 0.2 | 2.2 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.2 | 11.2 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 1.5 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.1 | 0.7 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 1.4 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 3.9 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 0.7 | GO:0051586 | peptidyl-cysteine methylation(GO:0018125) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.1 | 3.5 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 2.9 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.7 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.5 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 1.8 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 3.7 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.4 | GO:0051958 | methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.9 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.1 | 3.6 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.1 | 1.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.6 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.1 | 0.6 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 | 2.0 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 1.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 2.1 | GO:0007567 | parturition(GO:0007567) |
| 0.1 | 0.5 | GO:2001034 | chromatin silencing at telomere(GO:0006348) positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 0.9 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) cellular response to steroid hormone stimulus(GO:0071383) |
| 0.1 | 1.0 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.1 | 2.3 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.1 | 0.5 | GO:0061101 | neuroendocrine cell differentiation(GO:0061101) |
| 0.1 | 0.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.2 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.1 | 0.6 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.1 | 4.4 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 1.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.8 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.1 | 2.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 1.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 1.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 3.4 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 6.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.8 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 0.6 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 6.7 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 2.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 3.9 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 1.6 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 1.2 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.0 | 2.6 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 1.3 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) peptidyl-tyrosine modification(GO:0018212) |
| 0.0 | 3.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.7 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.9 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.5 | GO:0006631 | fatty acid metabolic process(GO:0006631) |
| 0.0 | 0.9 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.6 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 1.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 4.9 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 1.0 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.5 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.9 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.2 | GO:0015865 | purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.2 | GO:0016101 | retinoid metabolic process(GO:0001523) diterpenoid metabolic process(GO:0016101) |
| 0.0 | 2.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.2 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 1.7 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.1 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.7 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.6 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.1 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 60.5 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 4.0 | 19.8 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 2.4 | 7.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 2.1 | 22.6 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 2.0 | 8.0 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 1.9 | 11.6 | GO:1990357 | terminal web(GO:1990357) |
| 1.9 | 33.9 | GO:0034709 | methylosome(GO:0034709) |
| 1.8 | 10.7 | GO:0071817 | MMXD complex(GO:0071817) |
| 1.7 | 5.2 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 1.6 | 6.2 | GO:0000811 | GINS complex(GO:0000811) |
| 1.4 | 11.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 1.2 | 3.6 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 1.0 | 4.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 1.0 | 18.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.9 | 6.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.9 | 6.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.8 | 14.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.8 | 8.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.8 | 4.7 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.8 | 13.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.7 | 5.6 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.7 | 8.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.6 | 3.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.6 | 5.6 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.6 | 2.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.6 | 6.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.6 | 5.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.5 | 12.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.5 | 3.7 | GO:0070522 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.5 | 5.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.5 | 9.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.5 | 7.0 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.5 | 2.5 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.5 | 2.0 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.5 | 7.4 | GO:0090543 | Flemming body(GO:0090543) |
| 0.4 | 4.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.4 | 5.5 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.4 | 3.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.4 | 9.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.4 | 13.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.4 | 2.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.4 | 2.5 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.3 | 1.7 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.3 | 2.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.3 | 18.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.3 | 9.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.3 | 5.4 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.3 | 13.3 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.3 | 3.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 6.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.2 | 3.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 20.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 4.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 0.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.2 | 1.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.2 | 3.9 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.2 | 2.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.2 | 3.1 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.2 | 6.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 2.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.2 | 0.7 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.2 | 3.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 3.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 3.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 3.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 3.0 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 4.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 5.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 2.0 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.9 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 2.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 4.3 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 7.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 8.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 3.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 1.9 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 0.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 2.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 0.6 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 3.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 3.5 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 7.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 3.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.3 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 1.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 2.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 3.9 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.8 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 2.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 3.1 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 9.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 2.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 3.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 9.9 | GO:0005774 | vacuolar membrane(GO:0005774) |
| 0.0 | 2.1 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 2.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 4.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.3 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 23.4 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 2.3 | 7.0 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 2.0 | 12.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 1.8 | 5.4 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 1.5 | 7.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 1.4 | 26.8 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 1.3 | 4.0 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 1.3 | 8.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 1.3 | 5.1 | GO:0008184 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 1.2 | 3.6 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 1.2 | 3.5 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 1.1 | 6.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 1.0 | 10.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.9 | 2.6 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.9 | 12.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.7 | 4.4 | GO:0047115 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 0.7 | 5.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.7 | 6.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.7 | 4.7 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.7 | 2.0 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.7 | 13.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.6 | 5.8 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.6 | 6.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.6 | 5.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.6 | 1.8 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.6 | 18.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.6 | 2.9 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.6 | 2.3 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.6 | 1.7 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.6 | 3.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.5 | 2.6 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.5 | 3.7 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.5 | 4.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.5 | 23.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.5 | 2.9 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.5 | 6.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.4 | 12.9 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.4 | 6.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.4 | 9.6 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.4 | 3.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.4 | 2.1 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.4 | 2.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.4 | 3.6 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.4 | 4.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.4 | 6.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.4 | 6.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.4 | 3.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.3 | 8.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 1.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.3 | 8.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.3 | 0.9 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.3 | 3.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.3 | 4.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.3 | 3.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.3 | 5.8 | GO:0097493 | muscle alpha-actinin binding(GO:0051371) structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 3.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 1.4 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.2 | 2.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 2.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 2.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 4.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.2 | 0.6 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) coenzyme transporter activity(GO:0051185) |
| 0.2 | 5.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 5.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 1.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 3.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.2 | 2.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 2.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 7.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 4.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 4.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.8 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 2.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 9.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 3.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.9 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.6 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 2.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 2.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 6.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.4 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.1 | 6.5 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 4.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 3.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 2.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 23.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 9.3 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 0.4 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 11.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 8.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 2.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.5 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.1 | 7.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 3.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 4.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.6 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.3 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 1.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 12.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 1.7 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 2.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 17.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.6 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 2.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.5 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 4.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 3.1 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 2.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.6 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 2.8 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 2.4 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 4.1 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.9 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 4.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) Ran GTPase binding(GO:0008536) |
| 0.0 | 3.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.5 | GO:0060089 | receptor activity(GO:0004872) molecular transducer activity(GO:0060089) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 16.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.5 | 16.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.5 | 17.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.4 | 12.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.3 | 12.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.3 | 9.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 7.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 6.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 14.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 3.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 15.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 4.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.2 | 9.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 36.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.2 | 15.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 6.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 3.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 4.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 3.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 3.2 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 2.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 1.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 4.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 4.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.7 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 4.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 2.0 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 1.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 6.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 4.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 1.5 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 3.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 2.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 18.2 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 1.0 | 19.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.6 | 11.8 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.6 | 8.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.6 | 15.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.6 | 16.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.4 | 18.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.4 | 5.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.4 | 9.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.4 | 5.9 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.4 | 6.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.4 | 6.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.4 | 6.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.3 | 9.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 7.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.3 | 4.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 13.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.3 | 6.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.3 | 42.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.3 | 9.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 5.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 3.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 5.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 5.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 3.8 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.2 | 6.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 11.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.2 | 5.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 3.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 4.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 3.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 3.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 3.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 3.1 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 3.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 0.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 3.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 3.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 3.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 3.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 4.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 7.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 6.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 6.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 4.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.9 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 8.3 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 5.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 4.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.8 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 1.1 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 2.3 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 3.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |