Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for TCF7L2
Z-value: 0.81
Transcription factors associated with TCF7L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF7L2
|
ENSG00000148737.11 | transcription factor 7 like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF7L2 | hg19_v2_chr10_+_114709999_114710031 | 0.46 | 6.4e-13 | Click! |
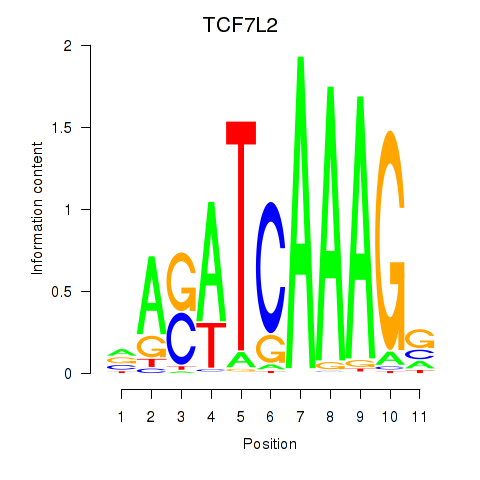
Activity profile of TCF7L2 motif
Sorted Z-values of TCF7L2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_69455855 | 21.60 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr5_-_16936340 | 17.13 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chr11_+_114128522 | 16.30 |
ENST00000535401.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr17_-_40169659 | 12.31 |
ENST00000457167.4
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chrX_-_10588459 | 11.46 |
ENST00000380782.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chrX_-_10588595 | 10.63 |
ENST00000423614.1
ENST00000317552.4 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr13_+_76334795 | 10.48 |
ENST00000526202.1
ENST00000465261.2 |
LMO7
|
LIM domain 7 |
| chr10_+_54074033 | 10.44 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr3_-_134093275 | 10.38 |
ENST00000513145.1
ENST00000422605.2 |
AMOTL2
|
angiomotin like 2 |
| chr13_+_76334567 | 9.85 |
ENST00000321797.8
|
LMO7
|
LIM domain 7 |
| chr3_-_134093395 | 9.53 |
ENST00000249883.5
|
AMOTL2
|
angiomotin like 2 |
| chr4_-_157892498 | 9.16 |
ENST00000502773.1
|
PDGFC
|
platelet derived growth factor C |
| chr1_-_94079648 | 9.16 |
ENST00000370247.3
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr10_+_114709999 | 8.78 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr3_-_149375783 | 8.71 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr2_+_234621551 | 8.58 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr5_+_102201509 | 8.10 |
ENST00000348126.2
ENST00000379787.4 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr10_+_114710211 | 7.99 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr5_-_175815565 | 7.43 |
ENST00000509257.1
ENST00000507413.1 ENST00000510123.1 |
NOP16
|
NOP16 nucleolar protein |
| chr7_+_73868439 | 6.86 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr7_-_27213893 | 6.47 |
ENST00000283921.4
|
HOXA10
|
homeobox A10 |
| chr1_-_54303934 | 6.10 |
ENST00000537333.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr4_-_139163491 | 5.95 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr1_-_54304212 | 5.90 |
ENST00000540001.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr5_+_102201430 | 5.60 |
ENST00000438793.3
ENST00000346918.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr1_-_54303949 | 5.15 |
ENST00000234725.8
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr6_+_32146131 | 5.15 |
ENST00000375094.3
|
RNF5
|
ring finger protein 5, E3 ubiquitin protein ligase |
| chr1_-_110933663 | 5.13 |
ENST00000369781.4
ENST00000541986.1 ENST00000369779.4 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr10_+_105005644 | 4.90 |
ENST00000441178.2
|
RP11-332O19.5
|
ribulose-5-phosphate-3-epimerase-like 1 |
| chr1_-_110933611 | 4.82 |
ENST00000472422.2
ENST00000437429.2 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr5_-_16742330 | 4.75 |
ENST00000505695.1
ENST00000427430.2 |
MYO10
|
myosin X |
| chr10_+_71561630 | 4.70 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr2_-_9770706 | 4.68 |
ENST00000381844.4
|
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
| chr17_-_56065484 | 4.62 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr5_-_148930960 | 4.57 |
ENST00000261798.5
ENST00000377843.2 |
CSNK1A1
|
casein kinase 1, alpha 1 |
| chrX_+_54835493 | 4.55 |
ENST00000396224.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr14_-_65409502 | 4.52 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr2_-_9771075 | 4.44 |
ENST00000446619.1
ENST00000238081.3 |
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
| chr17_+_34848049 | 4.34 |
ENST00000588902.1
ENST00000591067.1 |
ZNHIT3
|
zinc finger, HIT-type containing 3 |
| chr1_+_147013182 | 4.33 |
ENST00000234739.3
|
BCL9
|
B-cell CLL/lymphoma 9 |
| chr10_+_71561649 | 4.31 |
ENST00000398978.3
ENST00000354547.3 ENST00000357811.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr5_+_131593364 | 4.29 |
ENST00000253754.3
ENST00000379018.3 |
PDLIM4
|
PDZ and LIM domain 4 |
| chr16_-_73082274 | 4.15 |
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr17_+_75447326 | 4.13 |
ENST00000591088.1
|
SEPT9
|
septin 9 |
| chr10_+_71561704 | 4.07 |
ENST00000520267.1
|
COL13A1
|
collagen, type XIII, alpha 1 |
| chr6_-_11779403 | 3.90 |
ENST00000414691.3
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr2_+_69240302 | 3.88 |
ENST00000303714.4
|
ANTXR1
|
anthrax toxin receptor 1 |
| chrX_-_47004878 | 3.88 |
ENST00000377811.3
|
NDUFB11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
| chr15_+_43985725 | 3.87 |
ENST00000413453.2
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chrX_-_47004437 | 3.87 |
ENST00000276062.8
|
NDUFB11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
| chr2_+_69240415 | 3.77 |
ENST00000409829.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr12_-_49582593 | 3.62 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr17_+_37618257 | 3.60 |
ENST00000447079.4
|
CDK12
|
cyclin-dependent kinase 12 |
| chr19_-_3061397 | 3.59 |
ENST00000586839.1
|
AES
|
amino-terminal enhancer of split |
| chr10_+_101542462 | 3.57 |
ENST00000370449.4
ENST00000370434.1 |
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr2_+_69240511 | 3.55 |
ENST00000409349.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr7_-_27219849 | 3.54 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr5_+_140797296 | 3.47 |
ENST00000398594.2
|
PCDHGB7
|
protocadherin gamma subfamily B, 7 |
| chr22_+_41347363 | 3.39 |
ENST00000216225.8
|
RBX1
|
ring-box 1, E3 ubiquitin protein ligase |
| chr6_+_64281906 | 3.33 |
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr1_+_110091189 | 3.32 |
ENST00000369851.4
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr7_-_148581251 | 3.20 |
ENST00000478654.1
ENST00000460911.1 ENST00000350995.2 |
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr13_+_102142296 | 3.10 |
ENST00000376162.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr6_-_11779174 | 2.98 |
ENST00000379413.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr11_-_108422926 | 2.83 |
ENST00000428840.1
ENST00000526312.1 |
EXPH5
|
exophilin 5 |
| chr3_-_113465065 | 2.83 |
ENST00000497255.1
ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr7_-_148581360 | 2.75 |
ENST00000320356.2
ENST00000541220.1 ENST00000483967.1 ENST00000536783.1 |
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr15_+_57511609 | 2.75 |
ENST00000543579.1
ENST00000537840.1 ENST00000343827.3 |
TCF12
|
transcription factor 12 |
| chr21_-_35284635 | 2.74 |
ENST00000429238.1
|
AP000304.12
|
AP000304.12 |
| chr2_+_201936707 | 2.62 |
ENST00000433898.1
ENST00000454214.1 |
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr12_+_122326662 | 2.59 |
ENST00000261817.2
ENST00000538613.1 ENST00000542602.1 |
PSMD9
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr12_+_122326630 | 2.57 |
ENST00000541212.1
ENST00000340175.5 |
PSMD9
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr16_+_29819372 | 2.55 |
ENST00000568544.1
ENST00000569978.1 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr15_+_43885252 | 2.54 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chrX_-_153599578 | 2.51 |
ENST00000360319.4
ENST00000344736.4 |
FLNA
|
filamin A, alpha |
| chr1_-_109935819 | 2.50 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr15_+_43985084 | 2.46 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr6_-_32145861 | 2.41 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr17_-_39093672 | 2.38 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr2_-_88427568 | 2.30 |
ENST00000393750.3
ENST00000295834.3 |
FABP1
|
fatty acid binding protein 1, liver |
| chr7_-_148725733 | 2.30 |
ENST00000286091.4
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chrX_-_43832711 | 2.25 |
ENST00000378062.5
|
NDP
|
Norrie disease (pseudoglioma) |
| chr6_-_11779840 | 2.22 |
ENST00000506810.1
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr9_-_27005686 | 2.15 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chr6_-_112575912 | 2.14 |
ENST00000522006.1
ENST00000230538.7 ENST00000519932.1 |
LAMA4
|
laminin, alpha 4 |
| chr8_-_16859690 | 2.09 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr19_-_39924349 | 2.02 |
ENST00000602153.1
|
RPS16
|
ribosomal protein S16 |
| chr12_+_1738363 | 1.99 |
ENST00000397196.2
|
WNT5B
|
wingless-type MMTV integration site family, member 5B |
| chr5_+_170846640 | 1.95 |
ENST00000274625.5
|
FGF18
|
fibroblast growth factor 18 |
| chr1_-_153522562 | 1.87 |
ENST00000368714.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr17_-_46035187 | 1.81 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr5_+_140734570 | 1.80 |
ENST00000571252.1
|
PCDHGA4
|
protocadherin gamma subfamily A, 4 |
| chr2_+_228678550 | 1.72 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr8_+_32579341 | 1.68 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr10_-_104192405 | 1.64 |
ENST00000369937.4
|
CUEDC2
|
CUE domain containing 2 |
| chr12_-_49582978 | 1.60 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chrX_-_39956656 | 1.57 |
ENST00000397354.3
ENST00000378444.4 |
BCOR
|
BCL6 corepressor |
| chr10_+_115312766 | 1.56 |
ENST00000351270.3
|
HABP2
|
hyaluronan binding protein 2 |
| chr17_+_68100989 | 1.55 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr5_-_135290705 | 1.54 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr4_+_30721968 | 1.51 |
ENST00000361762.2
|
PCDH7
|
protocadherin 7 |
| chr16_+_29819446 | 1.50 |
ENST00000568282.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr2_+_71357434 | 1.49 |
ENST00000244230.2
|
MPHOSPH10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr15_-_31393910 | 1.39 |
ENST00000397795.2
ENST00000256552.6 ENST00000559179.1 |
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr5_-_157002775 | 1.32 |
ENST00000257527.4
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr2_+_201936458 | 1.29 |
ENST00000237889.4
|
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chrX_+_100646190 | 1.24 |
ENST00000471855.1
|
RPL36A
|
ribosomal protein L36a |
| chr5_+_175815732 | 1.20 |
ENST00000274787.2
|
HIGD2A
|
HIG1 hypoxia inducible domain family, member 2A |
| chr17_-_73389737 | 1.19 |
ENST00000392563.1
|
GRB2
|
growth factor receptor-bound protein 2 |
| chr3_+_141106643 | 1.13 |
ENST00000514251.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr6_-_112575687 | 1.13 |
ENST00000521398.1
ENST00000424408.2 ENST00000243219.3 |
LAMA4
|
laminin, alpha 4 |
| chr16_+_72088376 | 1.12 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr12_+_85673868 | 1.09 |
ENST00000316824.3
|
ALX1
|
ALX homeobox 1 |
| chr17_+_42923686 | 1.05 |
ENST00000591513.1
|
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr17_-_7297519 | 1.02 |
ENST00000576362.1
ENST00000571078.1 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr14_+_23235886 | 1.01 |
ENST00000604262.1
ENST00000431881.2 ENST00000412791.1 ENST00000358043.5 |
OXA1L
|
oxidase (cytochrome c) assembly 1-like |
| chr8_-_119964434 | 0.96 |
ENST00000297350.4
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b |
| chr14_+_75230011 | 0.96 |
ENST00000552421.1
ENST00000325680.7 ENST00000238571.3 |
YLPM1
|
YLP motif containing 1 |
| chrX_+_100645812 | 0.95 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr1_+_16693578 | 0.83 |
ENST00000401088.4
ENST00000471507.1 ENST00000401089.3 ENST00000375590.3 ENST00000492354.1 |
SZRD1
|
SUZ RNA binding domain containing 1 |
| chr6_-_112575838 | 0.83 |
ENST00000455073.1
|
LAMA4
|
laminin, alpha 4 |
| chr1_+_154244987 | 0.82 |
ENST00000328703.7
ENST00000457918.2 ENST00000483970.2 ENST00000435087.1 ENST00000532105.1 |
HAX1
|
HCLS1 associated protein X-1 |
| chrX_+_15525426 | 0.77 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr1_+_62417957 | 0.76 |
ENST00000307297.7
ENST00000543708.1 |
INADL
|
InaD-like (Drosophila) |
| chr3_+_29323043 | 0.73 |
ENST00000452462.1
ENST00000456853.1 |
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr5_+_112074029 | 0.73 |
ENST00000512211.2
|
APC
|
adenomatous polyposis coli |
| chr14_+_21236586 | 0.70 |
ENST00000326783.3
|
EDDM3B
|
epididymal protein 3B |
| chr7_-_80141328 | 0.66 |
ENST00000398291.3
|
GNAT3
|
guanine nucleotide binding protein, alpha transducing 3 |
| chr17_+_79679299 | 0.65 |
ENST00000331531.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr10_-_105677427 | 0.56 |
ENST00000369764.1
|
OBFC1
|
oligonucleotide/oligosaccharide-binding fold containing 1 |
| chrX_+_47004639 | 0.52 |
ENST00000345781.6
|
RBM10
|
RNA binding motif protein 10 |
| chr7_-_75452673 | 0.51 |
ENST00000416943.1
|
CCL24
|
chemokine (C-C motif) ligand 24 |
| chr8_+_29953163 | 0.49 |
ENST00000518192.1
|
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chr12_-_52779433 | 0.47 |
ENST00000257951.3
|
KRT84
|
keratin 84 |
| chr19_-_40791302 | 0.45 |
ENST00000392038.2
ENST00000578123.1 |
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr6_+_152130240 | 0.45 |
ENST00000427531.2
|
ESR1
|
estrogen receptor 1 |
| chrX_+_47004599 | 0.43 |
ENST00000329236.7
|
RBM10
|
RNA binding motif protein 10 |
| chr20_-_17511962 | 0.43 |
ENST00000377873.3
|
BFSP1
|
beaded filament structural protein 1, filensin |
| chr6_-_112575758 | 0.40 |
ENST00000431543.2
ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4
|
laminin, alpha 4 |
| chr14_+_32798547 | 0.28 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr22_+_19950060 | 0.28 |
ENST00000449653.1
|
COMT
|
catechol-O-methyltransferase |
| chrX_+_69488155 | 0.28 |
ENST00000374495.3
|
ARR3
|
arrestin 3, retinal (X-arrestin) |
| chrX_+_152760397 | 0.27 |
ENST00000331595.4
ENST00000431891.1 |
BGN
|
biglycan |
| chrX_-_63005405 | 0.25 |
ENST00000374878.1
ENST00000437457.2 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr8_-_133123406 | 0.24 |
ENST00000434736.2
|
HHLA1
|
HERV-H LTR-associating 1 |
| chr6_+_29427548 | 0.23 |
ENST00000377132.1
|
OR2H1
|
olfactory receptor, family 2, subfamily H, member 1 |
| chr17_-_38256973 | 0.18 |
ENST00000246672.3
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr17_-_7297833 | 0.18 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr5_-_58335281 | 0.17 |
ENST00000358923.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr12_+_52056548 | 0.16 |
ENST00000545061.1
ENST00000355133.3 |
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr7_-_16505440 | 0.14 |
ENST00000307068.4
|
SOSTDC1
|
sclerostin domain containing 1 |
| chr5_+_175288631 | 0.12 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr1_-_32801825 | 0.10 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr1_+_151138526 | 0.07 |
ENST00000368902.1
|
SCNM1
|
sodium channel modifier 1 |
| chr14_-_75179774 | 0.06 |
ENST00000555249.1
ENST00000556202.1 ENST00000356357.4 ENST00000338772.5 |
AREL1
AC007956.1
|
apoptosis resistant E3 ubiquitin protein ligase 1 Full-length cDNA 5-PRIME end of clone CS0CAP004YO05 of Thymus of Homo sapiens (human); Uncharacterized protein |
| chr8_-_95220775 | 0.03 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr17_-_39646116 | 0.01 |
ENST00000328119.6
|
KRT36
|
keratin 36 |
| chr2_-_55647057 | 0.00 |
ENST00000436346.1
|
CCDC88A
|
coiled-coil domain containing 88A |
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF7L2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.2 | GO:0030474 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 3.7 | 11.2 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 3.5 | 10.4 | GO:0009996 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) negative regulation of cell fate specification(GO:0009996) Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 3.4 | 13.7 | GO:0031179 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 2.5 | 15.3 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 2.2 | 21.6 | GO:0070141 | response to UV-A(GO:0070141) |
| 2.0 | 6.0 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 1.7 | 8.6 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 1.7 | 22.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 1.4 | 6.9 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 1.4 | 16.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 1.2 | 3.6 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) response to antineoplastic agent(GO:0097327) |
| 1.0 | 4.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 1.0 | 5.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.9 | 12.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.8 | 8.7 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.8 | 5.3 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.7 | 2.1 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.7 | 4.6 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.6 | 2.5 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.6 | 6.3 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.6 | 1.7 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.6 | 2.3 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.5 | 2.8 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.4 | 10.8 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.4 | 2.5 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.4 | 2.0 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.4 | 18.0 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.4 | 1.1 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.4 | 9.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.4 | 10.0 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.3 | 3.4 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.3 | 1.0 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.3 | 1.6 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.3 | 3.6 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.3 | 5.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.3 | 21.9 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.3 | 6.0 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.3 | 13.1 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.2 | 2.5 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.2 | 2.4 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 | 0.7 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.2 | 2.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.2 | 0.6 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.2 | 9.1 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.2 | 4.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.2 | 11.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 3.6 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.2 | 2.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.2 | 2.8 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 2.5 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.1 | 0.8 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 7.4 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 0.5 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 4.3 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.1 | 1.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 3.3 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.1 | 4.8 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.1 | 0.2 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.1 | 0.3 | GO:0045963 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.1 | 0.8 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 2.0 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 0.8 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.5 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 2.8 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 19.6 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 0.2 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.5 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 2.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 5.1 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 1.4 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 3.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.5 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 2.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 2.2 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 2.4 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.0 | 1.9 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.7 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 1.1 | GO:0001843 | neural tube closure(GO:0001843) tube closure(GO:0060606) |
| 0.0 | 1.8 | GO:0070268 | cornification(GO:0070268) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.2 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 3.3 | 13.1 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 2.5 | 15.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 1.1 | 3.4 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 1.1 | 21.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.8 | 2.5 | GO:0031523 | Myb complex(GO:0031523) |
| 0.5 | 3.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.5 | 5.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.5 | 8.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.4 | 6.0 | GO:0045120 | pronucleus(GO:0045120) |
| 0.4 | 5.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.4 | 2.8 | GO:0031415 | NatA complex(GO:0031415) |
| 0.4 | 25.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.4 | 27.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.3 | 1.4 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.3 | 11.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.3 | 2.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.3 | 1.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 5.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.2 | 4.1 | GO:0031105 | septin complex(GO:0031105) |
| 0.2 | 11.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 3.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.8 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 18.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 4.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 1.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 2.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 6.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 4.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 10.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 17.7 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 1.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 2.2 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 7.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 4.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.6 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 10.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 10.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 4.8 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 1.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 9.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.1 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 16.3 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 3.4 | 13.7 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 2.4 | 21.9 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 1.2 | 6.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.9 | 10.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.8 | 6.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.7 | 21.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.7 | 13.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.7 | 6.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.6 | 2.5 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.6 | 17.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.6 | 1.7 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.5 | 2.5 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.5 | 2.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.4 | 5.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.4 | 3.4 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.4 | 9.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.4 | 12.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.3 | 2.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 9.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.3 | 3.6 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.3 | 3.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 8.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 2.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 0.6 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.2 | 1.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 21.7 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.2 | 10.0 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.2 | 1.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 3.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 2.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.5 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 11.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 2.3 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 4.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 4.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 1.9 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 9.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 11.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 1.9 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 2.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 2.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 8.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 3.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 3.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 13.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.8 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 0.5 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 1.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 4.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 12.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 4.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.3 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 1.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 1.0 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.6 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 10.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 1.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 4.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 5.5 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 21.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.5 | 11.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.4 | 21.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.3 | 10.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 10.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 13.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 3.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 7.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 9.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 10.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 3.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 4.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 5.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 15.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 3.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 3.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 6.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 0.8 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 19.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 2.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 28.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.9 | 21.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.5 | 9.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 21.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.3 | 8.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 4.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.2 | 5.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 13.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 3.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.2 | 5.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 16.6 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 6.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 3.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 3.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 5.2 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.1 | 2.5 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 3.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.5 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.0 | 9.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 2.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 2.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 2.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |