Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
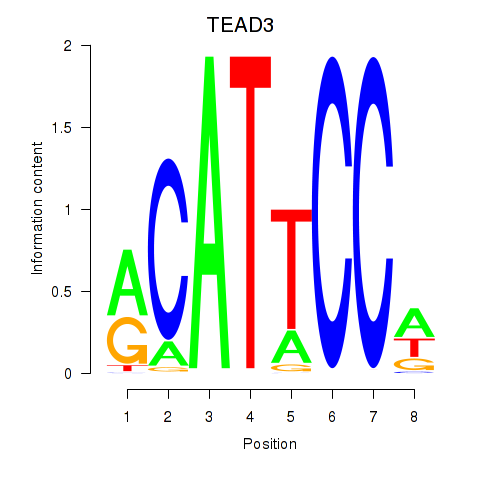
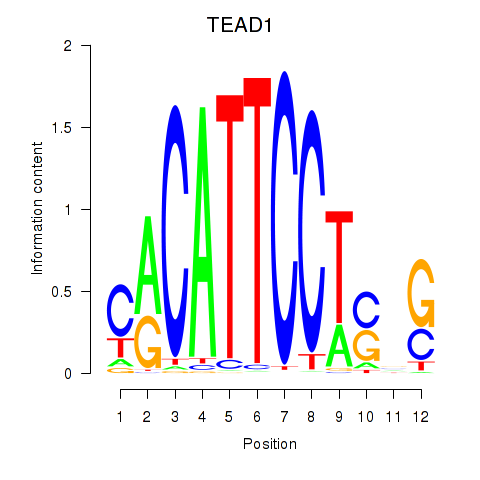
Results for TEAD3_TEAD1
Z-value: 2.74
Transcription factors associated with TEAD3_TEAD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TEAD3
|
ENSG00000007866.14 | TEA domain transcription factor 3 |
|
TEAD1
|
ENSG00000187079.10 | TEA domain transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TEAD3 | hg19_v2_chr6_-_35464727_35464738, hg19_v2_chr6_-_35464817_35464894 | 0.58 | 3.8e-21 | Click! |
| TEAD1 | hg19_v2_chr11_+_12766583_12766592, hg19_v2_chr11_+_12696102_12696164, hg19_v2_chr11_+_12695944_12695989 | 0.53 | 2.3e-17 | Click! |
Activity profile of TEAD3_TEAD1 motif
Sorted Z-values of TEAD3_TEAD1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_132272504 | 208.59 |
ENST00000367976.3
|
CTGF
|
connective tissue growth factor |
| chr10_-_90712520 | 124.85 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr1_+_86046433 | 81.33 |
ENST00000451137.2
|
CYR61
|
cysteine-rich, angiogenic inducer, 61 |
| chr11_-_111781454 | 64.46 |
ENST00000533280.1
|
CRYAB
|
crystallin, alpha B |
| chr7_+_134464414 | 60.30 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr5_-_39425068 | 55.00 |
ENST00000515700.1
ENST00000339788.6 |
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr17_-_40575535 | 54.30 |
ENST00000357037.5
|
PTRF
|
polymerase I and transcript release factor |
| chr1_-_95391315 | 54.17 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr15_+_63334831 | 53.57 |
ENST00000288398.6
ENST00000358278.3 ENST00000560445.1 ENST00000403994.3 ENST00000357980.4 ENST00000559556.1 ENST00000559397.1 ENST00000267996.7 ENST00000560970.1 |
TPM1
|
tropomyosin 1 (alpha) |
| chr2_-_216257849 | 50.57 |
ENST00000456923.1
|
FN1
|
fibronectin 1 |
| chr4_+_184020398 | 49.41 |
ENST00000403733.3
ENST00000378925.3 |
WWC2
|
WW and C2 domain containing 2 |
| chr7_+_116165754 | 48.02 |
ENST00000405348.1
|
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr12_-_7245125 | 47.99 |
ENST00000542285.1
ENST00000540610.1 |
C1R
|
complement component 1, r subcomponent |
| chr20_+_35169885 | 41.58 |
ENST00000279022.2
ENST00000346786.2 |
MYL9
|
myosin, light chain 9, regulatory |
| chr15_+_39873268 | 40.99 |
ENST00000397591.2
ENST00000260356.5 |
THBS1
|
thrombospondin 1 |
| chr7_+_134464376 | 40.78 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr7_+_128470431 | 40.78 |
ENST00000325888.8
ENST00000346177.6 |
FLNC
|
filamin C, gamma |
| chr15_+_96869165 | 38.36 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr6_+_30850697 | 38.17 |
ENST00000509639.1
ENST00000412274.2 ENST00000507901.1 ENST00000507046.1 ENST00000437124.2 ENST00000454612.2 ENST00000396342.2 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr8_+_144816303 | 37.45 |
ENST00000533004.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr1_-_1293904 | 36.48 |
ENST00000309212.6
ENST00000342753.4 ENST00000445648.2 |
MXRA8
|
matrix-remodelling associated 8 |
| chr11_-_111782696 | 36.03 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr6_+_83073334 | 35.20 |
ENST00000369750.3
|
TPBG
|
trophoblast glycoprotein |
| chr5_-_39425222 | 35.05 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr7_+_116165038 | 34.97 |
ENST00000393470.1
|
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr6_+_83072923 | 34.90 |
ENST00000535040.1
|
TPBG
|
trophoblast glycoprotein |
| chr11_-_111781610 | 34.60 |
ENST00000525823.1
|
CRYAB
|
crystallin, alpha B |
| chr5_-_39425290 | 34.50 |
ENST00000545653.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr17_+_20059302 | 32.70 |
ENST00000395530.2
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr11_-_111782484 | 32.43 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr3_-_134093275 | 31.78 |
ENST00000513145.1
ENST00000422605.2 |
AMOTL2
|
angiomotin like 2 |
| chr7_+_100464760 | 31.46 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr4_-_187644930 | 31.32 |
ENST00000441802.2
|
FAT1
|
FAT atypical cadherin 1 |
| chr11_+_114166536 | 31.30 |
ENST00000299964.3
|
NNMT
|
nicotinamide N-methyltransferase |
| chr10_-_33247124 | 31.28 |
ENST00000414670.1
ENST00000302278.3 ENST00000374956.4 ENST00000488494.1 ENST00000417122.2 ENST00000474568.1 |
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
| chr11_-_111781554 | 31.17 |
ENST00000526167.1
ENST00000528961.1 |
CRYAB
|
crystallin, alpha B |
| chr15_-_60690163 | 31.03 |
ENST00000558998.1
ENST00000560165.1 ENST00000557986.1 ENST00000559780.1 ENST00000559467.1 ENST00000559956.1 ENST00000332680.4 ENST00000396024.3 ENST00000421017.2 ENST00000560466.1 ENST00000558132.1 ENST00000559113.1 ENST00000557906.1 ENST00000558558.1 ENST00000560468.1 ENST00000559370.1 ENST00000558169.1 ENST00000559725.1 ENST00000558985.1 ENST00000451270.2 |
ANXA2
|
annexin A2 |
| chr3_-_134093395 | 30.15 |
ENST00000249883.5
|
AMOTL2
|
angiomotin like 2 |
| chr3_-_114343039 | 29.72 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr10_-_99447024 | 27.32 |
ENST00000370626.3
|
AVPI1
|
arginine vasopressin-induced 1 |
| chr5_-_55290773 | 26.96 |
ENST00000502326.3
ENST00000381298.2 |
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor) |
| chr1_-_201438282 | 26.86 |
ENST00000367311.3
ENST00000367309.1 |
PHLDA3
|
pleckstrin homology-like domain, family A, member 3 |
| chrX_-_10588459 | 26.71 |
ENST00000380782.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr1_-_94050668 | 24.97 |
ENST00000539242.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr10_+_123872483 | 23.65 |
ENST00000369001.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr1_-_16344500 | 23.49 |
ENST00000406363.2
ENST00000411503.1 ENST00000545268.1 ENST00000487046.1 |
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular) |
| chr11_-_14379997 | 23.47 |
ENST00000526063.1
ENST00000532814.1 |
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr3_-_127541194 | 23.42 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase |
| chr7_+_134430212 | 22.51 |
ENST00000436461.2
|
CALD1
|
caldesmon 1 |
| chr1_-_161279749 | 22.46 |
ENST00000533357.1
ENST00000360451.6 ENST00000336559.4 |
MPZ
|
myelin protein zero |
| chr4_-_186733363 | 22.13 |
ENST00000393523.2
ENST00000393528.3 ENST00000449407.2 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr16_-_122619 | 21.77 |
ENST00000262316.6
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila) |
| chr11_+_92085707 | 21.27 |
ENST00000525166.1
|
FAT3
|
FAT atypical cadherin 3 |
| chr6_-_128841503 | 21.18 |
ENST00000368215.3
ENST00000532331.1 ENST00000368213.5 ENST00000368207.3 ENST00000525459.1 ENST00000368210.3 ENST00000368226.4 ENST00000368227.3 |
PTPRK
|
protein tyrosine phosphatase, receptor type, K |
| chr19_-_14606900 | 20.18 |
ENST00000393029.3
ENST00000393028.1 ENST00000393033.4 ENST00000345425.2 ENST00000586027.1 ENST00000591349.1 ENST00000587210.1 |
GIPC1
|
GIPC PDZ domain containing family, member 1 |
| chr11_+_92577506 | 19.65 |
ENST00000533797.1
|
FAT3
|
FAT atypical cadherin 3 |
| chr19_+_33182823 | 18.65 |
ENST00000397061.3
|
NUDT19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
| chr12_-_8815299 | 18.42 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr11_+_66824276 | 18.36 |
ENST00000308831.2
|
RHOD
|
ras homolog family member D |
| chr19_+_45409011 | 18.18 |
ENST00000252486.4
ENST00000446996.1 ENST00000434152.1 |
APOE
|
apolipoprotein E |
| chr1_-_16482554 | 18.00 |
ENST00000358432.5
|
EPHA2
|
EPH receptor A2 |
| chr12_-_8815215 | 17.77 |
ENST00000544889.1
ENST00000543369.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr11_+_92085262 | 17.51 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr3_-_128712955 | 17.48 |
ENST00000265068.5
|
KIAA1257
|
KIAA1257 |
| chr15_-_48937982 | 17.29 |
ENST00000316623.5
|
FBN1
|
fibrillin 1 |
| chr4_+_41614909 | 17.09 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr19_-_17185848 | 16.59 |
ENST00000593360.1
|
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr2_-_190044480 | 16.53 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr14_-_23904861 | 16.49 |
ENST00000355349.3
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta |
| chr6_-_35464817 | 16.21 |
ENST00000338863.7
|
TEAD3
|
TEA domain family member 3 |
| chr6_-_35464727 | 15.81 |
ENST00000402886.3
|
TEAD3
|
TEA domain family member 3 |
| chr4_+_41614720 | 15.43 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr17_+_4901199 | 15.08 |
ENST00000320785.5
ENST00000574165.1 |
KIF1C
|
kinesin family member 1C |
| chr2_-_217560248 | 14.83 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr6_-_30712313 | 14.58 |
ENST00000376377.2
ENST00000259874.5 |
IER3
|
immediate early response 3 |
| chr4_+_169552748 | 14.45 |
ENST00000504519.1
ENST00000512127.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr1_-_59043166 | 14.40 |
ENST00000371225.2
|
TACSTD2
|
tumor-associated calcium signal transducer 2 |
| chr20_-_44540686 | 14.30 |
ENST00000477313.1
ENST00000542937.1 ENST00000372431.3 ENST00000354050.4 ENST00000420868.2 |
PLTP
|
phospholipid transfer protein |
| chr7_-_137686791 | 14.01 |
ENST00000452463.1
ENST00000330387.6 ENST00000456390.1 |
CREB3L2
|
cAMP responsive element binding protein 3-like 2 |
| chr3_+_147127142 | 13.99 |
ENST00000282928.4
|
ZIC1
|
Zic family member 1 |
| chrX_-_10851762 | 13.94 |
ENST00000380785.1
ENST00000380787.1 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr11_-_8832182 | 13.90 |
ENST00000527510.1
ENST00000528527.1 ENST00000528523.1 ENST00000313726.6 |
ST5
|
suppression of tumorigenicity 5 |
| chr19_+_15218180 | 13.81 |
ENST00000342784.2
ENST00000597977.1 ENST00000600440.1 |
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr18_+_21693306 | 13.40 |
ENST00000540918.2
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr7_+_102715315 | 13.22 |
ENST00000428183.2
ENST00000323716.3 ENST00000441711.2 ENST00000454559.1 ENST00000425331.1 ENST00000541300.1 |
ARMC10
|
armadillo repeat containing 10 |
| chr1_+_196621002 | 13.14 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr8_+_70378852 | 12.98 |
ENST00000525061.1
ENST00000458141.2 ENST00000260128.4 |
SULF1
|
sulfatase 1 |
| chr11_-_76381029 | 12.68 |
ENST00000407242.2
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr8_-_82395461 | 12.58 |
ENST00000256104.4
|
FABP4
|
fatty acid binding protein 4, adipocyte |
| chr17_+_37856299 | 12.53 |
ENST00000269571.5
|
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr3_-_185538849 | 12.49 |
ENST00000421047.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr9_-_14314518 | 12.40 |
ENST00000397581.2
|
NFIB
|
nuclear factor I/B |
| chr9_-_14314566 | 12.24 |
ENST00000397579.2
|
NFIB
|
nuclear factor I/B |
| chr17_-_39677971 | 12.23 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr19_-_50143452 | 12.22 |
ENST00000246792.3
|
RRAS
|
related RAS viral (r-ras) oncogene homolog |
| chr9_+_118916082 | 12.06 |
ENST00000328252.3
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr12_-_8815404 | 11.77 |
ENST00000359478.2
ENST00000396549.2 |
MFAP5
|
microfibrillar associated protein 5 |
| chr7_+_26438187 | 11.63 |
ENST00000439120.1
ENST00000430548.1 ENST00000421862.1 ENST00000449537.1 ENST00000420774.1 ENST00000418758.2 |
AC004540.5
|
AC004540.5 |
| chr2_+_189157536 | 11.53 |
ENST00000409580.1
ENST00000409637.3 |
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr6_+_134210243 | 11.51 |
ENST00000367882.4
|
TCF21
|
transcription factor 21 |
| chr2_+_102759199 | 11.43 |
ENST00000409288.1
ENST00000410023.1 |
IL1R1
|
interleukin 1 receptor, type I |
| chr2_+_189157498 | 11.36 |
ENST00000359135.3
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr3_-_48057890 | 11.33 |
ENST00000434267.1
|
MAP4
|
microtubule-associated protein 4 |
| chr5_-_43412418 | 11.15 |
ENST00000537013.1
ENST00000361115.4 |
CCL28
|
chemokine (C-C motif) ligand 28 |
| chr1_+_150229554 | 10.93 |
ENST00000369111.4
|
CA14
|
carbonic anhydrase XIV |
| chr12_-_54813229 | 10.74 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr1_-_229569834 | 10.73 |
ENST00000366684.3
ENST00000366683.2 |
ACTA1
|
actin, alpha 1, skeletal muscle |
| chr12_+_27677085 | 10.71 |
ENST00000545334.1
ENST00000540114.1 ENST00000537927.1 ENST00000318304.8 ENST00000535047.1 ENST00000542629.1 ENST00000228425.6 |
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chr1_+_196621156 | 10.66 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr15_+_63354769 | 10.61 |
ENST00000558910.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr16_+_30387141 | 10.42 |
ENST00000566955.1
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr4_-_186877806 | 10.32 |
ENST00000355634.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_-_149375783 | 10.18 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr17_+_37856253 | 10.06 |
ENST00000540147.1
ENST00000584450.1 |
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr5_+_140868717 | 9.98 |
ENST00000252087.1
|
PCDHGC5
|
protocadherin gamma subfamily C, 5 |
| chr10_-_33625154 | 9.93 |
ENST00000265371.4
|
NRP1
|
neuropilin 1 |
| chr13_-_33760216 | 9.86 |
ENST00000255486.4
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr17_-_73505961 | 9.81 |
ENST00000433559.2
|
CASKIN2
|
CASK interacting protein 2 |
| chr12_-_111358372 | 9.71 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr11_+_46299199 | 9.68 |
ENST00000529193.1
ENST00000288400.3 |
CREB3L1
|
cAMP responsive element binding protein 3-like 1 |
| chr4_+_176986978 | 9.67 |
ENST00000508596.1
ENST00000393643.2 |
WDR17
|
WD repeat domain 17 |
| chr8_+_97597148 | 9.60 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr4_-_186877502 | 9.46 |
ENST00000431902.1
ENST00000284776.7 ENST00000415274.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_-_94079648 | 9.29 |
ENST00000370247.3
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr1_+_114522049 | 9.24 |
ENST00000369551.1
ENST00000320334.4 |
OLFML3
|
olfactomedin-like 3 |
| chr9_-_16727978 | 9.14 |
ENST00000418777.1
ENST00000468187.2 |
BNC2
|
basonuclin 2 |
| chr17_-_73511584 | 9.10 |
ENST00000321617.3
|
CASKIN2
|
CASK interacting protein 2 |
| chr11_-_111170526 | 9.10 |
ENST00000355430.4
|
COLCA1
|
colorectal cancer associated 1 |
| chr18_+_3247779 | 8.89 |
ENST00000578611.1
ENST00000583449.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr10_-_17659234 | 8.83 |
ENST00000466335.1
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr18_-_25616519 | 8.76 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr17_-_39093672 | 8.58 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr1_+_62417957 | 8.57 |
ENST00000307297.7
ENST00000543708.1 |
INADL
|
InaD-like (Drosophila) |
| chr1_-_11918988 | 8.50 |
ENST00000376468.3
|
NPPB
|
natriuretic peptide B |
| chr1_-_201915590 | 8.48 |
ENST00000367288.4
|
LMOD1
|
leiomodin 1 (smooth muscle) |
| chr8_-_38325219 | 8.35 |
ENST00000533668.1
ENST00000413133.2 ENST00000397108.4 ENST00000526742.1 ENST00000525001.1 ENST00000425967.3 ENST00000529552.1 ENST00000397113.2 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr17_+_57232690 | 8.04 |
ENST00000262293.4
|
PRR11
|
proline rich 11 |
| chr18_+_3247413 | 7.96 |
ENST00000579226.1
ENST00000217652.3 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr17_+_70117153 | 7.89 |
ENST00000245479.2
|
SOX9
|
SRY (sex determining region Y)-box 9 |
| chr6_+_118869452 | 7.85 |
ENST00000357525.5
|
PLN
|
phospholamban |
| chr16_+_30386098 | 7.79 |
ENST00000322861.7
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr5_-_150138551 | 7.70 |
ENST00000446090.2
ENST00000447998.2 |
DCTN4
|
dynactin 4 (p62) |
| chr15_+_33010175 | 7.65 |
ENST00000300177.4
ENST00000560677.1 ENST00000560830.1 |
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr6_+_148663729 | 7.64 |
ENST00000367467.3
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr2_+_30569506 | 7.57 |
ENST00000421976.2
|
AC109642.1
|
AC109642.1 |
| chr6_-_3157760 | 7.54 |
ENST00000333628.3
|
TUBB2A
|
tubulin, beta 2A class IIa |
| chr19_+_39138320 | 7.53 |
ENST00000424234.2
ENST00000390009.3 ENST00000589528.1 |
ACTN4
|
actinin, alpha 4 |
| chr8_-_119124045 | 7.52 |
ENST00000378204.2
|
EXT1
|
exostosin glycosyltransferase 1 |
| chr17_+_73717551 | 7.44 |
ENST00000450894.3
|
ITGB4
|
integrin, beta 4 |
| chr9_-_113341985 | 7.31 |
ENST00000374469.1
|
SVEP1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr9_-_14308004 | 7.28 |
ENST00000493697.1
|
NFIB
|
nuclear factor I/B |
| chr17_+_73717407 | 7.28 |
ENST00000579662.1
|
ITGB4
|
integrin, beta 4 |
| chr15_+_74466744 | 7.27 |
ENST00000560862.1
ENST00000395118.1 |
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr4_-_186732048 | 7.16 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr12_+_52445191 | 7.06 |
ENST00000243050.1
ENST00000394825.1 ENST00000550763.1 ENST00000394824.2 ENST00000548232.1 ENST00000562373.1 |
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr11_-_47470703 | 7.00 |
ENST00000298854.2
|
RAPSN
|
receptor-associated protein of the synapse |
| chr11_+_111782934 | 7.00 |
ENST00000304298.3
|
HSPB2
|
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr11_+_46316677 | 6.98 |
ENST00000534787.1
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1 |
| chr19_+_39138271 | 6.96 |
ENST00000252699.2
|
ACTN4
|
actinin, alpha 4 |
| chr19_+_676385 | 6.95 |
ENST00000166139.4
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein) |
| chr7_+_30960915 | 6.85 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr2_-_211179883 | 6.82 |
ENST00000352451.3
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr11_-_47470682 | 6.80 |
ENST00000529341.1
ENST00000352508.3 |
RAPSN
|
receptor-associated protein of the synapse |
| chr18_+_7754957 | 6.76 |
ENST00000400053.4
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr9_+_139873264 | 6.66 |
ENST00000446677.1
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain) |
| chr4_+_166300084 | 6.62 |
ENST00000402744.4
|
CPE
|
carboxypeptidase E |
| chr15_+_41136216 | 6.49 |
ENST00000562057.1
ENST00000344051.4 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr9_-_13175823 | 6.48 |
ENST00000545857.1
|
MPDZ
|
multiple PDZ domain protein |
| chr15_+_41136586 | 6.38 |
ENST00000431806.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr4_+_165675269 | 6.35 |
ENST00000507311.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chrX_-_41449204 | 6.18 |
ENST00000378179.3
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr12_+_32655048 | 6.09 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr7_-_135412925 | 6.08 |
ENST00000354042.4
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr9_+_112542572 | 6.04 |
ENST00000374530.3
|
PALM2-AKAP2
|
PALM2-AKAP2 readthrough |
| chr8_-_141728760 | 6.03 |
ENST00000430260.2
|
PTK2
|
protein tyrosine kinase 2 |
| chr12_-_104443890 | 6.01 |
ENST00000547583.1
ENST00000360814.4 ENST00000546851.1 |
GLT8D2
|
glycosyltransferase 8 domain containing 2 |
| chr3_+_38347427 | 5.96 |
ENST00000273173.4
|
SLC22A14
|
solute carrier family 22, member 14 |
| chr11_+_101981169 | 5.95 |
ENST00000526343.1
ENST00000282441.5 ENST00000537274.1 ENST00000345877.2 |
YAP1
|
Yes-associated protein 1 |
| chr11_-_47470591 | 5.91 |
ENST00000524487.1
|
RAPSN
|
receptor-associated protein of the synapse |
| chr10_+_123923205 | 5.87 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr19_+_782755 | 5.86 |
ENST00000606242.1
ENST00000586061.1 |
AC006273.5
|
AC006273.5 |
| chr14_+_32546485 | 5.78 |
ENST00000345122.3
ENST00000432921.1 ENST00000433497.1 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr4_-_175443943 | 5.77 |
ENST00000296522.6
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr9_+_131314859 | 5.76 |
ENST00000358161.5
ENST00000372731.4 ENST00000372739.3 |
SPTAN1
|
spectrin, alpha, non-erythrocytic 1 |
| chr4_+_25657444 | 5.75 |
ENST00000504570.1
ENST00000382051.3 |
SLC34A2
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 2 |
| chr1_+_119957554 | 5.68 |
ENST00000543831.1
ENST00000433745.1 ENST00000369416.3 |
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chrX_-_135333514 | 5.66 |
ENST00000370661.1
ENST00000370660.3 |
MAP7D3
|
MAP7 domain containing 3 |
| chr14_-_89021077 | 5.58 |
ENST00000556564.1
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr14_+_74035763 | 5.57 |
ENST00000238651.5
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr1_+_26503894 | 5.57 |
ENST00000361530.6
ENST00000374253.5 |
CNKSR1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr19_-_49371711 | 5.53 |
ENST00000355496.5
ENST00000263265.6 |
PLEKHA4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr12_+_3068466 | 5.51 |
ENST00000358409.2
|
TEAD4
|
TEA domain family member 4 |
| chr7_-_83278322 | 5.51 |
ENST00000307792.3
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr8_+_120079478 | 5.48 |
ENST00000332843.2
|
COLEC10
|
collectin sub-family member 10 (C-type lectin) |
| chr2_+_32502952 | 5.43 |
ENST00000238831.4
|
YIPF4
|
Yip1 domain family, member 4 |
| chr1_+_100818484 | 5.42 |
ENST00000544534.1
|
CDC14A
|
cell division cycle 14A |
| chr14_-_23877474 | 5.38 |
ENST00000405093.3
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr1_+_152974218 | 5.37 |
ENST00000331860.3
ENST00000443178.1 ENST00000295367.4 |
SPRR3
|
small proline-rich protein 3 |
| chr5_+_54455946 | 5.34 |
ENST00000503787.1
ENST00000296734.6 ENST00000515370.1 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr10_+_104005272 | 5.30 |
ENST00000369983.3
|
GBF1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TEAD3_TEAD1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 69.5 | 208.6 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 31.1 | 124.5 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 27.7 | 83.0 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 22.6 | 135.6 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 16.9 | 50.6 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 13.7 | 41.0 | GO:0002605 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) negative regulation of dendritic cell antigen processing and presentation(GO:0002605) |
| 11.0 | 198.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 10.4 | 31.3 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 10.3 | 31.0 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 9.6 | 38.4 | GO:0009956 | radial pattern formation(GO:0009956) |
| 9.1 | 18.2 | GO:0032803 | regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) |
| 9.0 | 81.3 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) chondroblast differentiation(GO:0060591) |
| 9.0 | 18.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 8.4 | 33.7 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 8.0 | 64.2 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 7.9 | 7.9 | GO:0072197 | ureter urothelium development(GO:0072190) ureter morphogenesis(GO:0072197) |
| 6.6 | 19.7 | GO:1900073 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 6.4 | 25.6 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 6.3 | 31.5 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 5.4 | 27.1 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 5.3 | 32.0 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 4.9 | 14.8 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 4.8 | 14.5 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 4.6 | 31.9 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 4.4 | 22.0 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 4.2 | 38.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 4.1 | 16.5 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 3.8 | 11.5 | GO:0072276 | branchiomeric skeletal muscle development(GO:0014707) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 3.8 | 11.4 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 3.6 | 14.3 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 3.5 | 17.3 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 3.5 | 13.8 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 3.3 | 23.4 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 3.1 | 40.6 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 2.9 | 66.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 2.8 | 8.4 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 2.8 | 8.4 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 2.8 | 16.7 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 2.6 | 31.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 2.5 | 15.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 2.3 | 4.6 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 2.3 | 6.9 | GO:0072019 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) metanephric proximal tubule development(GO:0072237) |
| 2.2 | 15.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 2.2 | 11.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 2.2 | 6.6 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 2.2 | 11.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 2.2 | 13.0 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 2.1 | 36.5 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 2.1 | 87.7 | GO:0035329 | hippo signaling(GO:0035329) |
| 2.1 | 12.7 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 2.0 | 4.0 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 2.0 | 2.0 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 2.0 | 9.9 | GO:0060301 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 2.0 | 9.9 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 2.0 | 11.8 | GO:1901631 | positive regulation of presynaptic membrane organization(GO:1901631) |
| 2.0 | 27.4 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 1.9 | 7.6 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 1.8 | 3.6 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 1.8 | 3.6 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 1.8 | 60.7 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 1.6 | 34.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 1.6 | 4.8 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.6 | 3.2 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 1.5 | 49.1 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 1.4 | 8.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 1.4 | 6.9 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 1.4 | 9.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 1.4 | 4.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 1.3 | 4.0 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 1.3 | 5.3 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 1.3 | 6.6 | GO:0030070 | insulin processing(GO:0030070) |
| 1.3 | 8.9 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 1.3 | 3.8 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 1.3 | 2.5 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 1.2 | 9.7 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 1.2 | 4.6 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 1.1 | 4.6 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 1.1 | 3.3 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 1.1 | 3.2 | GO:0060214 | endocardium formation(GO:0060214) |
| 1.1 | 14.8 | GO:0035878 | nail development(GO:0035878) |
| 1.0 | 5.2 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 1.0 | 23.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.0 | 3.0 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 1.0 | 10.7 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 1.0 | 2.9 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 1.0 | 1.9 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 1.0 | 1.9 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 1.0 | 3.8 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.9 | 7.5 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.9 | 9.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.9 | 8.4 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.9 | 70.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.9 | 3.7 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.9 | 3.6 | GO:0072660 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.9 | 8.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.9 | 5.4 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.9 | 3.5 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.8 | 8.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.8 | 3.4 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.8 | 3.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.8 | 3.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.8 | 46.3 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.8 | 2.5 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.8 | 18.5 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.8 | 12.3 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.7 | 20.2 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.7 | 14.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.7 | 5.8 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.7 | 12.9 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.7 | 4.3 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.7 | 2.8 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.7 | 5.6 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.7 | 4.8 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.7 | 2.0 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.7 | 103.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.7 | 6.0 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.7 | 9.9 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.6 | 56.8 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.6 | 10.0 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.6 | 1.9 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.6 | 1.8 | GO:0006726 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.6 | 1.2 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.6 | 1.7 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.5 | 2.7 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.5 | 2.1 | GO:0023021 | glucosylceramide catabolic process(GO:0006680) termination of signal transduction(GO:0023021) beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.5 | 2.1 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.5 | 3.6 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.5 | 10.3 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.5 | 3.0 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.5 | 9.8 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.5 | 12.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.5 | 4.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.5 | 20.7 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.5 | 1.4 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.5 | 1.8 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.5 | 2.7 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.5 | 5.0 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.5 | 2.3 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.4 | 4.0 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.4 | 6.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.4 | 3.5 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.4 | 5.7 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.4 | 9.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.4 | 48.0 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.4 | 5.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.4 | 2.5 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.4 | 3.5 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.4 | 3.5 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.4 | 8.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.4 | 2.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.4 | 26.2 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.4 | 3.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 1.7 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.3 | 27.4 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.3 | 3.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.3 | 2.0 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.3 | 1.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.3 | 1.9 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.3 | 3.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 2.6 | GO:1905247 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.3 | 25.9 | GO:0070268 | cornification(GO:0070268) |
| 0.3 | 117.9 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.3 | 12.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.3 | 2.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.3 | 3.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.3 | 1.5 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.2 | 1.7 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 | 1.5 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.2 | 0.7 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.2 | 0.7 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.2 | 3.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 2.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 21.8 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.2 | 26.0 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.2 | 1.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.2 | 0.9 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.2 | 5.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 4.5 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.2 | 4.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.2 | 13.5 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.2 | 2.6 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.2 | 2.6 | GO:2000394 | phosphatidylcholine catabolic process(GO:0034638) positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.2 | 21.7 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.2 | 23.5 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.2 | 1.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.2 | 0.5 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.2 | 27.2 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.2 | 15.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.2 | 16.8 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.2 | 0.5 | GO:0045112 | integrin biosynthetic process(GO:0045112) regulation of integrin biosynthetic process(GO:0045113) positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.2 | 1.0 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.2 | 2.2 | GO:0030220 | platelet formation(GO:0030220) |
| 0.2 | 2.5 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.2 | 12.3 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.2 | 4.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 1.9 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.2 | 5.7 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.2 | 5.6 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.2 | 31.4 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.2 | 6.2 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 0.7 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 1.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 2.4 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.1 | 0.5 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 5.1 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 2.5 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.1 | 4.0 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 0.8 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 11.5 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.1 | 1.3 | GO:0042346 | positive regulation of NF-kappaB import into nucleus(GO:0042346) |
| 0.1 | 5.1 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.1 | 1.6 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.1 | 1.2 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.1 | 1.8 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 10.7 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.1 | 0.8 | GO:0098719 | gastric acid secretion(GO:0001696) sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 1.2 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 4.0 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.8 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.9 | GO:0061037 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) |
| 0.1 | 2.0 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 1.2 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 3.3 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.1 | 0.1 | GO:0070638 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.1 | 8.8 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.1 | 0.5 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 5.3 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.1 | 5.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.7 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 0.3 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.1 | 1.0 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 0.7 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 0.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 1.6 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.1 | 1.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 1.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.9 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 1.5 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 2.7 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 3.5 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 2.8 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 2.6 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 0.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.9 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.0 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 1.6 | GO:0030879 | mammary gland development(GO:0030879) |
| 0.0 | 0.1 | GO:0044254 | angiotensin catabolic process in blood(GO:0002005) multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) |
| 0.0 | 0.1 | GO:2000510 | positive regulation of sperm motility involved in capacitation(GO:0060474) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.7 | GO:0006479 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 0.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 2.9 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 31.2 | 124.9 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 13.8 | 124.5 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 13.5 | 215.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 10.4 | 31.3 | GO:0034680 | integrin alpha3-beta1 complex(GO:0034667) integrin alpha8-beta1 complex(GO:0034678) integrin alpha10-beta1 complex(GO:0034680) |
| 6.9 | 90.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 6.5 | 97.9 | GO:0030478 | actin cap(GO:0030478) |
| 6.3 | 31.5 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 6.2 | 31.0 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 6.2 | 24.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 6.1 | 18.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 5.7 | 80.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 5.5 | 16.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 3.8 | 64.2 | GO:0032059 | bleb(GO:0032059) |
| 3.6 | 213.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 3.3 | 72.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 2.7 | 10.7 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 2.7 | 31.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 2.5 | 89.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 2.3 | 6.9 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 2.2 | 19.7 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 2.0 | 9.9 | GO:0060091 | kinocilium(GO:0060091) |
| 1.8 | 16.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 1.7 | 3.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 1.6 | 14.8 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 1.6 | 4.7 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 1.5 | 4.6 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 1.4 | 4.3 | GO:0000806 | Y chromosome(GO:0000806) cyclin A2-CDK2 complex(GO:0097124) |
| 1.3 | 4.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 1.3 | 4.0 | GO:0031523 | Myb complex(GO:0031523) |
| 1.2 | 5.9 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 1.1 | 12.0 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 1.0 | 14.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.9 | 45.5 | GO:0043034 | costamere(GO:0043034) |
| 0.8 | 12.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.8 | 8.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.8 | 3.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.7 | 22.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.7 | 4.8 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.7 | 16.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.6 | 41.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.6 | 5.4 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.6 | 61.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.5 | 9.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.5 | 19.2 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.5 | 1.6 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.5 | 7.6 | GO:0043234 | protein complex(GO:0043234) |
| 0.5 | 4.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.5 | 7.8 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.4 | 3.6 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.4 | 179.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.4 | 2.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.4 | 3.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 2.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.4 | 36.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.4 | 75.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.4 | 20.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.4 | 6.2 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.4 | 126.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.4 | 27.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.3 | 14.5 | GO:0002102 | podosome(GO:0002102) |
| 0.3 | 13.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.3 | 29.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.3 | 15.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.3 | 2.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.3 | 2.0 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.3 | 2.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 15.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 11.2 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.3 | 0.5 | GO:0071437 | invadopodium(GO:0071437) invadopodium membrane(GO:0071438) |
| 0.2 | 20.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 21.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 17.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.2 | 3.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 2.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 1.8 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 1.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 12.4 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.2 | 6.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 6.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 1.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 5.0 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 3.5 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.2 | 10.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 0.9 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 3.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 1.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 5.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.7 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 6.2 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 1.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 4.5 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.1 | 3.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 6.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 11.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 92.8 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.1 | 76.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 1.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 11.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.3 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 2.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 6.9 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 10.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.3 | GO:0044215 | host(GO:0018995) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 1.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 27.7 | 83.0 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 13.7 | 41.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 11.2 | 33.7 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 10.9 | 261.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 7.8 | 31.3 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 7.6 | 38.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 6.4 | 196.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 6.1 | 18.2 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 5.8 | 127.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 5.2 | 31.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 4.9 | 136.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 3.2 | 13.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 3.1 | 96.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 2.9 | 46.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 2.9 | 11.4 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 2.7 | 26.9 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 2.6 | 18.4 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 2.5 | 20.3 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 2.5 | 10.0 | GO:0038132 | neuregulin binding(GO:0038132) |
| 2.5 | 27.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 2.5 | 54.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 2.3 | 6.9 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 2.0 | 7.8 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 1.9 | 5.7 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) steroid delta-isomerase activity(GO:0004769) |
| 1.9 | 7.5 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 1.9 | 1.9 | GO:0031432 | titin binding(GO:0031432) RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 1.8 | 23.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 1.8 | 46.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 1.8 | 3.5 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 1.7 | 6.7 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 1.6 | 31.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 1.5 | 24.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 1.5 | 29.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 1.5 | 30.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 1.4 | 5.8 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 1.4 | 36.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 1.4 | 8.4 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 1.3 | 4.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 1.3 | 26.6 | GO:0031005 | filamin binding(GO:0031005) |
| 1.3 | 14.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 1.2 | 96.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 1.2 | 8.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 1.2 | 3.5 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 1.2 | 16.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 1.1 | 9.9 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 1.1 | 3.3 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 1.1 | 3.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 1.1 | 5.4 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 1.0 | 19.7 | GO:0033130 | acetylcholine receptor binding(GO:0033130) protein anchor(GO:0043495) |
| 1.0 | 4.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 1.0 | 3.0 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 1.0 | 50.6 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 1.0 | 38.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 1.0 | 23.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.9 | 9.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.9 | 7.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.9 | 20.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.9 | 3.5 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.8 | 3.4 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.8 | 15.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.8 | 2.5 | GO:0015403 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.8 | 4.0 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.8 | 72.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.7 | 5.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.7 | 5.7 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.7 | 7.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.7 | 4.6 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.6 | 2.6 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.6 | 10.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.6 | 5.0 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.6 | 5.6 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.6 | 3.6 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.6 | 7.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.5 | 6.0 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.5 | 5.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.5 | 2.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.5 | 32.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.5 | 6.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.5 | 2.7 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.4 | 1.7 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.4 | 3.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.4 | 1.7 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.4 | 12.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.4 | 2.4 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.4 | 34.7 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.3 | 1.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.3 | 8.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.3 | 2.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.3 | 2.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.3 | 5.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.3 | 32.9 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.3 | 6.8 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.3 | 1.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.3 | 3.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.3 | 34.4 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.3 | 5.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.3 | 3.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.3 | 5.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 3.0 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.3 | 4.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.3 | 0.5 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.3 | 2.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 2.0 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.2 | 36.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.2 | 10.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 11.2 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.2 | 7.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 22.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 9.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 175.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.2 | 12.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 0.9 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 5.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 4.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 5.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 11.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 2.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 12.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 0.8 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 5.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 5.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 13.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 0.5 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.2 | 1.9 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 2.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 4.3 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.2 | 2.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 12.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.7 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 2.8 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 41.8 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 6.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 31.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 3.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 17.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 13.7 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.1 | 3.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 11.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 3.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 4.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 1.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 5.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 0.7 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 0.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.8 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.1 | 0.8 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 3.9 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 2.1 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 13.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 4.8 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 2.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 7.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 2.1 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 2.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.3 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 2.8 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 10.4 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.1 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 1.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.6 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.9 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 6.6 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 2.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 4.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 2.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 2.1 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.2 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 1.4 | GO:0047485 | protein N-terminus binding(GO:0047485) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 149.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 4.1 | 397.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 2.0 | 116.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 1.4 | 23.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 1.1 | 39.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 1.1 | 4.6 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.9 | 21.7 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.9 | 12.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.9 | 42.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.8 | 30.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.8 | 63.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.7 | 25.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.7 | 40.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.7 | 56.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.6 | 9.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.6 | 115.6 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.6 | 26.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.6 | 6.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.5 | 9.9 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.5 | 17.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.5 | 43.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.4 | 8.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.4 | 16.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.4 | 11.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.4 | 18.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.3 | 8.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.3 | 4.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.3 | 49.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 4.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.3 | 5.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.2 | 9.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 7.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 5.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 13.3 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.2 | 4.0 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 7.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 6.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 5.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 4.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.2 | 4.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.2 | 5.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.2 | 31.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 1.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 7.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 1.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 0.6 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 17.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 2.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 15.0 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 2.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 265.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 6.9 | 124.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 5.7 | 362.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 4.1 | 119.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 3.5 | 38.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 2.5 | 61.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 2.3 | 64.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 2.3 | 55.0 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 1.5 | 31.9 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 1.3 | 32.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 1.3 | 23.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 1.3 | 37.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 1.2 | 22.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 1.0 | 26.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.9 | 23.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.8 | 20.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.7 | 10.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.7 | 25.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.7 | 17.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.5 | 26.6 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.5 | 5.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.5 | 11.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.5 | 7.4 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.5 | 6.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.4 | 15.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.4 | 5.7 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.4 | 44.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.4 | 9.9 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.3 | 8.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.3 | 4.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.3 | 8.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.3 | 7.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 32.6 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.3 | 8.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 6.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.3 | 16.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 10.7 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.2 | 13.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 6.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.2 | 11.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.2 | 2.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 3.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 15.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 9.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 4.9 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 0.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 5.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 4.3 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.1 | 6.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 2.9 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.1 | 1.2 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.1 | 9.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 5.2 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 2.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.4 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.1 | 0.9 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 2.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 1.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 3.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |