Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
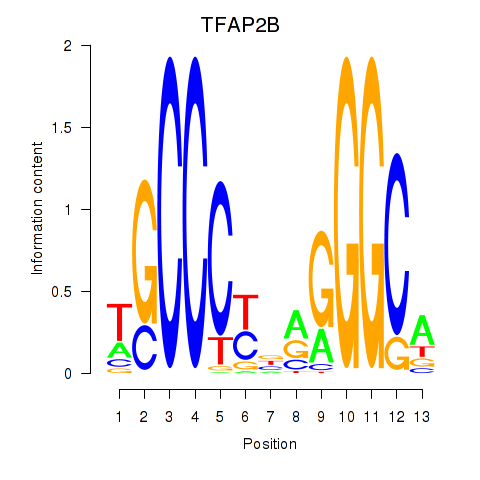
Results for TFAP2B
Z-value: 1.16
Transcription factors associated with TFAP2B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP2B
|
ENSG00000008196.8 | transcription factor AP-2 beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFAP2B | hg19_v2_chr6_+_50786414_50786439 | 0.33 | 6.3e-07 | Click! |
Activity profile of TFAP2B motif
Sorted Z-values of TFAP2B motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TFAP2B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.0 | 31.9 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 4.3 | 21.4 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 3.6 | 10.8 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 2.9 | 11.8 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 2.7 | 8.0 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 2.4 | 9.6 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 2.4 | 9.4 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 2.3 | 7.0 | GO:0002818 | intracellular defense response(GO:0002818) |
| 2.1 | 6.3 | GO:0070666 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 2.1 | 6.2 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 2.1 | 28.9 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 2.0 | 6.0 | GO:0003383 | apical constriction(GO:0003383) mesoderm migration involved in gastrulation(GO:0007509) |
| 1.9 | 13.5 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 1.7 | 5.1 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 1.5 | 6.0 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 1.4 | 5.6 | GO:1905174 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 1.4 | 4.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 1.4 | 4.1 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 1.3 | 18.8 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 1.3 | 5.3 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 1.3 | 9.2 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 1.3 | 5.3 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 1.3 | 6.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 1.2 | 3.7 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 1.1 | 6.9 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 1.1 | 4.5 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 1.0 | 5.0 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 1.0 | 15.7 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 1.0 | 2.9 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.9 | 2.8 | GO:0046732 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.9 | 3.7 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.9 | 3.7 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.9 | 4.6 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.9 | 6.4 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.9 | 4.5 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.9 | 5.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.9 | 2.6 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.8 | 3.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.8 | 3.3 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
| 0.8 | 8.1 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.8 | 2.4 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.8 | 13.3 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.8 | 14.8 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.8 | 2.3 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.8 | 6.2 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.8 | 2.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.8 | 3.8 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.7 | 11.9 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.7 | 2.2 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.7 | 3.5 | GO:0072313 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.7 | 1.4 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.7 | 7.0 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.7 | 10.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.6 | 6.5 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.6 | 2.5 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.6 | 1.3 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.6 | 3.8 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.6 | 1.8 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.6 | 1.8 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.6 | 1.8 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.6 | 2.9 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.6 | 2.3 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.6 | 11.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.6 | 1.7 | GO:0010193 | response to ozone(GO:0010193) |
| 0.6 | 1.7 | GO:0035283 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.6 | 5.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.6 | 1.7 | GO:0045659 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.6 | 3.9 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.6 | 1.7 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.5 | 1.6 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.5 | 48.4 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.5 | 3.1 | GO:0071504 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.5 | 2.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.5 | 3.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.5 | 1.8 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.5 | 1.8 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.5 | 13.8 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.4 | 4.0 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.4 | 2.6 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.4 | 1.7 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.4 | 2.6 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.4 | 1.3 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.4 | 0.4 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.4 | 0.8 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.4 | 3.7 | GO:0042278 | purine nucleoside metabolic process(GO:0042278) |
| 0.4 | 2.4 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.4 | 1.2 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.4 | 3.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.4 | 5.2 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.4 | 7.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.4 | 2.9 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.4 | 2.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.4 | 1.8 | GO:0090625 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.3 | 4.5 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.3 | 1.4 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.3 | 1.0 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.3 | 36.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.3 | 1.7 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.3 | 1.3 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.3 | 8.9 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.3 | 3.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 9.8 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.3 | 2.6 | GO:0060751 | branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.3 | 3.2 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.3 | 1.3 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.3 | 4.4 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.3 | 1.9 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.3 | 3.0 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to photoreceptor outer segment(GO:1903546) |
| 0.3 | 12.0 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.3 | 2.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.3 | 3.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.3 | 1.2 | GO:0060996 | dendritic spine development(GO:0060996) |
| 0.3 | 4.8 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.3 | 0.8 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) beta-alanine metabolic process(GO:0019482) |
| 0.3 | 1.4 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.3 | 0.8 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.3 | 0.8 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) primary lung bud formation(GO:0060431) lung induction(GO:0060492) |
| 0.3 | 1.1 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.3 | 1.3 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.3 | 1.0 | GO:0042414 | epinephrine metabolic process(GO:0042414) epinephrine biosynthetic process(GO:0042418) |
| 0.3 | 2.6 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.3 | 5.6 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.3 | 1.8 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.3 | 0.8 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.2 | 7.0 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.2 | 2.5 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.2 | 5.9 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.2 | 1.2 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.2 | 0.9 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.2 | 1.6 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 7.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.2 | 0.9 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.2 | 3.0 | GO:0001553 | luteinization(GO:0001553) |
| 0.2 | 16.7 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 4.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 0.9 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.2 | 4.4 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.2 | 1.7 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.2 | 10.1 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.2 | 4.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 2.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.2 | 3.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 2.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.2 | 1.0 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.2 | 1.0 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 0.8 | GO:0072299 | visceral serous pericardium development(GO:0061032) posterior mesonephric tubule development(GO:0072166) negative regulation of metanephric glomerulus development(GO:0072299) regulation of metanephric glomerular mesangial cell proliferation(GO:0072301) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.2 | 10.5 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.2 | 1.0 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.2 | 1.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.2 | 3.6 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.2 | 0.6 | GO:1905244 | fasciculation of motor neuron axon(GO:0097156) regulation of modification of synaptic structure(GO:1905244) |
| 0.2 | 0.7 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 1.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) positive regulation of t-circle formation(GO:1904431) |
| 0.2 | 2.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.2 | 1.3 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.2 | 3.6 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.2 | 3.5 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 | 2.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 7.4 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.6 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.1 | 1.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 2.0 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 | 0.8 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 4.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 2.5 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.1 | 9.2 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.1 | 0.4 | GO:0072423 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.1 | 1.3 | GO:0042416 | dopamine biosynthetic process(GO:0042416) |
| 0.1 | 3.5 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 3.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.3 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 2.6 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 1.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 2.9 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 2.2 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.1 | 5.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 7.0 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.1 | 6.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 1.5 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 3.3 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 1.4 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 1.4 | GO:0032196 | transposition(GO:0032196) |
| 0.1 | 7.3 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.1 | 2.7 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.1 | 0.4 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) multi-organism behavior(GO:0051705) |
| 0.1 | 1.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 3.2 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 3.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 2.1 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 | 0.4 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 0.7 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 1.6 | GO:0060070 | canonical Wnt signaling pathway(GO:0060070) |
| 0.1 | 7.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 1.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 1.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 1.5 | GO:0071624 | positive regulation of granulocyte chemotaxis(GO:0071624) |
| 0.1 | 4.1 | GO:0043507 | positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 6.0 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 1.6 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 4.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 2.7 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.1 | 0.6 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.1 | 0.7 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 4.4 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.1 | 1.7 | GO:0038202 | TORC1 signaling(GO:0038202) |
| 0.1 | 2.7 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.1 | 7.6 | GO:0007187 | G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger(GO:0007187) |
| 0.1 | 4.1 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.1 | 5.0 | GO:0007127 | meiosis I(GO:0007127) |
| 0.1 | 0.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 3.9 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.1 | 3.3 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.1 | 1.1 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.1 | 1.8 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.5 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.9 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.9 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 3.4 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 | 6.5 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.0 | 1.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.5 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 1.2 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 1.6 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.4 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.0 | 3.8 | GO:0006023 | aminoglycan biosynthetic process(GO:0006023) glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 1.5 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.5 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 1.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 1.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.6 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.1 | GO:0043385 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.0 | 0.1 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.0 | 3.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.1 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 4.1 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 3.8 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 2.8 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 1.0 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.6 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 2.6 | GO:0048839 | inner ear development(GO:0048839) |
| 0.0 | 3.4 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 1.6 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.2 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 1.1 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 1.1 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 4.3 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.0 | 0.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 4.1 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 1.1 | GO:0046546 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 1.0 | GO:0007586 | digestion(GO:0007586) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 18.8 | GO:0044609 | DBIRD complex(GO:0044609) |
| 3.1 | 9.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 2.1 | 19.0 | GO:0044194 | cytolytic granule(GO:0044194) |
| 2.1 | 14.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 1.7 | 1.7 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 1.4 | 4.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 1.3 | 3.9 | GO:0031251 | PAN complex(GO:0031251) |
| 1.2 | 3.5 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 1.1 | 5.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.9 | 10.7 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.9 | 4.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.9 | 9.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.8 | 5.8 | GO:0060091 | kinocilium(GO:0060091) |
| 0.6 | 14.9 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.6 | 2.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.6 | 4.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.5 | 2.7 | GO:0032021 | NELF complex(GO:0032021) |
| 0.5 | 10.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.5 | 4.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.5 | 3.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.5 | 5.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.5 | 3.0 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.5 | 4.6 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.5 | 50.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.5 | 3.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.4 | 3.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.4 | 1.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.4 | 55.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.3 | 1.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.3 | 1.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.3 | 4.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 4.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.3 | 9.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.3 | 3.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.3 | 1.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.3 | 2.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 2.9 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.3 | 3.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.3 | 2.3 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.3 | 5.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 5.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 2.6 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 0.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 4.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 1.8 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.2 | 1.0 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.2 | 4.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 2.6 | GO:0071437 | invadopodium(GO:0071437) |
| 0.2 | 1.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 1.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.1 | 0.9 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 5.1 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 20.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 1.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 2.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 13.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.0 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.1 | 10.2 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 4.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 2.3 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 9.3 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 5.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 10.2 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 6.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 6.7 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 3.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.4 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.1 | 1.8 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.1 | 6.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.0 | GO:0070069 | endoplasmic reticulum chaperone complex(GO:0034663) cytochrome complex(GO:0070069) |
| 0.1 | 1.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.1 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 0.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.6 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 1.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 2.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 3.6 | GO:0097708 | cytoplasmic vesicle(GO:0031410) intracellular vesicle(GO:0097708) |
| 0.0 | 3.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 3.7 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 3.0 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 13.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 10.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 17.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.2 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.6 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 3.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 3.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 3.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.6 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 5.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.9 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 69.7 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 4.1 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 21.4 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 3.2 | 31.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 2.6 | 15.7 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 2.5 | 44.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 2.4 | 12.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 2.3 | 7.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 2.3 | 11.7 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 2.0 | 8.1 | GO:0035276 | ethanol binding(GO:0035276) |
| 1.9 | 9.4 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 1.7 | 5.0 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 1.6 | 8.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 1.4 | 4.3 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.4 | 4.2 | GO:0015039 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 1.4 | 4.1 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 1.3 | 6.7 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 1.3 | 6.6 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 1.2 | 16.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 1.1 | 9.8 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 1.1 | 10.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 1.0 | 6.2 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.9 | 3.8 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.9 | 2.8 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.9 | 6.3 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.8 | 13.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.8 | 3.2 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.8 | 8.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.7 | 2.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.7 | 10.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.7 | 1.3 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.6 | 2.6 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.6 | 6.5 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.6 | 3.8 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.6 | 1.3 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.6 | 3.8 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.6 | 4.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.6 | 3.6 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.6 | 1.8 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.6 | 2.9 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.6 | 2.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.6 | 5.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.6 | 1.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.5 | 7.0 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.5 | 1.6 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.5 | 6.9 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.5 | 6.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.5 | 6.0 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.5 | 13.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.4 | 5.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.4 | 1.3 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.4 | 1.3 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.4 | 2.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.4 | 2.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.4 | 1.6 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.4 | 1.6 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.4 | 3.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 3.5 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.4 | 5.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.4 | 4.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.4 | 20.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.4 | 3.3 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.4 | 3.7 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.4 | 3.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.4 | 1.8 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.3 | 1.0 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.3 | 5.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.3 | 1.4 | GO:0004803 | transposase activity(GO:0004803) |
| 0.3 | 9.1 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.3 | 2.6 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.3 | 3.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.3 | 10.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.3 | 4.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.3 | 0.8 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.3 | 6.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.3 | 6.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 3.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.3 | 9.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.3 | 1.9 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.3 | 5.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.3 | 3.9 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.3 | 6.0 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.2 | 4.9 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.2 | 4.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 5.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 4.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 1.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.2 | 2.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 1.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 0.9 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.2 | 2.4 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.2 | 3.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 20.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 0.6 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.2 | 0.8 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.2 | 2.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 3.5 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.2 | 0.7 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 3.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 1.8 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 36.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 3.3 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.2 | 3.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 5.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.2 | 1.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 1.5 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.8 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.6 | GO:0080084 | RNA polymerase III transcription factor binding(GO:0001025) RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) 5S rDNA binding(GO:0080084) |
| 0.1 | 0.9 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 4.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.8 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.7 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.6 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 4.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 1.0 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 7.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.9 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 1.7 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 1.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 1.8 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 2.2 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 1.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.4 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 2.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 2.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 1.8 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 3.8 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 6.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 2.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 4.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 8.8 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.4 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 2.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.9 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 1.0 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 1.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 2.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 6.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 1.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 1.4 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 1.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 1.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0017057 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 7.3 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 1.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 2.5 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 5.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 4.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 2.5 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 1.6 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.9 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 1.4 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.8 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 4.5 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.5 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.0 | 9.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 2.6 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 3.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 8.4 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.6 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 4.5 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.8 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 2.6 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.5 | GO:0042623 | ATPase activity, coupled(GO:0042623) |
| 0.0 | 1.4 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 16.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 1.0 | 49.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.5 | 21.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.5 | 40.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.4 | 6.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.4 | 3.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.3 | 29.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.3 | 6.5 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.3 | 14.6 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.3 | 8.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.2 | 12.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 4.5 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.2 | 3.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 6.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 4.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 10.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 9.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.2 | 3.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 8.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.2 | 4.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 4.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 5.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 5.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.7 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 9.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 2.6 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 3.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 1.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 3.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 5.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 4.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 3.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 6.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 7.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 0.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 0.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 2.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 1.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.4 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 2.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 1.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 4.8 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 3.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 13.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 2.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 2.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 3.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 6.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 31.6 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 1.2 | 51.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.9 | 10.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.8 | 5.7 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.6 | 11.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.6 | 10.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.6 | 7.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.4 | 71.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.4 | 8.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.4 | 4.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.4 | 6.0 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.3 | 4.4 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.3 | 8.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.3 | 50.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.3 | 9.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.3 | 5.9 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.3 | 1.8 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 8.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.2 | 10.1 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.2 | 21.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 5.8 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.2 | 9.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 3.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 3.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 3.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 6.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 2.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 4.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 7.0 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 5.3 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.1 | 3.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 4.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 4.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 3.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 3.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 1.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 2.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 11.4 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 1.7 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 2.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 4.6 | REACTOME SIGNALLING BY NGF | Genes involved in Signalling by NGF |
| 0.1 | 7.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 2.1 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.1 | 2.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.1 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.0 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 0.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 4.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 2.1 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 1.3 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 1.0 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 3.5 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.5 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 2.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.6 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.7 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.1 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 3.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.3 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.8 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |