Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
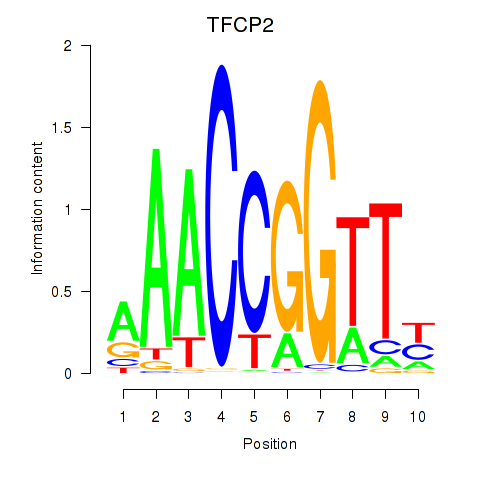
Results for TFCP2
Z-value: 1.16
Transcription factors associated with TFCP2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFCP2
|
ENSG00000135457.5 | transcription factor CP2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFCP2 | hg19_v2_chr12_-_51566562_51566584 | 0.18 | 7.7e-03 | Click! |
Activity profile of TFCP2 motif
Sorted Z-values of TFCP2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_40720974 | 35.24 |
ENST00000380748.1
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr21_-_40720995 | 33.18 |
ENST00000380749.5
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr4_-_174255536 | 31.62 |
ENST00000446922.2
|
HMGB2
|
high mobility group box 2 |
| chr2_-_61765315 | 30.92 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr17_-_5322786 | 26.04 |
ENST00000225696.4
|
NUP88
|
nucleoporin 88kDa |
| chr5_+_137514834 | 20.74 |
ENST00000508792.1
ENST00000504621.1 |
KIF20A
|
kinesin family member 20A |
| chr2_-_10952832 | 19.43 |
ENST00000540494.1
|
PDIA6
|
protein disulfide isomerase family A, member 6 |
| chr5_+_137514687 | 19.23 |
ENST00000394894.3
|
KIF20A
|
kinesin family member 20A |
| chr1_+_45212074 | 18.44 |
ENST00000372217.1
|
KIF2C
|
kinesin family member 2C |
| chr3_-_183966717 | 18.36 |
ENST00000446569.1
ENST00000418734.2 ENST00000397676.3 |
ALG3
|
ALG3, alpha-1,3- mannosyltransferase |
| chr9_+_75766652 | 18.31 |
ENST00000257497.6
|
ANXA1
|
annexin A1 |
| chr2_-_10952922 | 17.54 |
ENST00000272227.3
|
PDIA6
|
protein disulfide isomerase family A, member 6 |
| chrX_+_119737806 | 17.42 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr4_+_107236692 | 17.29 |
ENST00000510207.1
|
AIMP1
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chrX_+_118602363 | 17.22 |
ENST00000317881.8
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr15_-_34635314 | 15.67 |
ENST00000557912.1
ENST00000328848.4 |
NOP10
|
NOP10 ribonucleoprotein |
| chr6_+_138725343 | 15.17 |
ENST00000607197.1
ENST00000367697.3 |
HEBP2
|
heme binding protein 2 |
| chr1_+_45212051 | 14.97 |
ENST00000372222.3
|
KIF2C
|
kinesin family member 2C |
| chr14_-_23451467 | 14.95 |
ENST00000555074.1
ENST00000361265.4 |
RP11-298I3.5
AJUBA
|
RP11-298I3.5 ajuba LIM protein |
| chr7_-_7679633 | 14.58 |
ENST00000401447.1
|
RPA3
|
replication protein A3, 14kDa |
| chr7_-_105162652 | 14.37 |
ENST00000356362.2
ENST00000469408.1 |
PUS7
|
pseudouridylate synthase 7 homolog (S. cerevisiae) |
| chr6_+_142623063 | 13.52 |
ENST00000296932.8
ENST00000367609.3 |
GPR126
|
G protein-coupled receptor 126 |
| chrX_-_151938171 | 13.28 |
ENST00000393902.3
ENST00000417212.1 ENST00000370278.3 |
MAGEA3
|
melanoma antigen family A, 3 |
| chr10_-_95242044 | 13.17 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr1_-_54304212 | 12.85 |
ENST00000540001.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr10_-_95241951 | 12.56 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr2_+_200820269 | 12.38 |
ENST00000392290.1
|
C2orf47
|
chromosome 2 open reading frame 47 |
| chr3_-_113465065 | 12.35 |
ENST00000497255.1
ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr14_-_21737551 | 12.14 |
ENST00000554891.1
ENST00000555883.1 ENST00000553753.1 ENST00000555914.1 ENST00000557336.1 ENST00000555215.1 ENST00000556628.1 ENST00000555137.1 ENST00000556226.1 ENST00000555309.1 ENST00000556142.1 ENST00000554969.1 ENST00000554455.1 ENST00000556513.1 ENST00000557201.1 ENST00000420743.2 ENST00000557768.1 ENST00000553300.1 ENST00000554383.1 ENST00000554539.1 |
HNRNPC
|
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
| chrX_+_151867214 | 11.94 |
ENST00000329342.5
ENST00000412733.1 ENST00000457643.1 |
MAGEA6
|
melanoma antigen family A, 6 |
| chrX_-_151903184 | 11.84 |
ENST00000357916.4
ENST00000393869.3 |
MAGEA12
|
melanoma antigen family A, 12 |
| chrX_-_151903101 | 11.71 |
ENST00000393900.3
|
MAGEA12
|
melanoma antigen family A, 12 |
| chr6_+_3000057 | 11.69 |
ENST00000397717.2
|
NQO2
|
NAD(P)H dehydrogenase, quinone 2 |
| chr14_-_71107921 | 11.57 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr1_-_11118896 | 11.38 |
ENST00000465788.1
|
SRM
|
spermidine synthase |
| chr4_-_99850243 | 11.07 |
ENST00000280892.6
ENST00000511644.1 ENST00000504432.1 ENST00000505992.1 |
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr7_-_44613494 | 10.92 |
ENST00000431640.1
ENST00000258772.5 |
DDX56
|
DEAD (Asp-Glu-Ala-Asp) box helicase 56 |
| chr14_-_21737610 | 10.77 |
ENST00000320084.7
ENST00000449098.1 ENST00000336053.6 |
HNRNPC
|
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
| chr9_-_130213652 | 10.68 |
ENST00000536368.1
ENST00000361436.5 |
RPL12
|
ribosomal protein L12 |
| chr4_-_157892498 | 10.63 |
ENST00000502773.1
|
PDGFC
|
platelet derived growth factor C |
| chr17_-_40169659 | 10.55 |
ENST00000457167.4
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr1_-_229644034 | 10.55 |
ENST00000366678.3
ENST00000261396.3 ENST00000537506.1 |
NUP133
|
nucleoporin 133kDa |
| chr6_+_106959718 | 10.34 |
ENST00000369066.3
|
AIM1
|
absent in melanoma 1 |
| chr5_+_34757309 | 10.32 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr10_+_75936444 | 10.30 |
ENST00000372734.3
ENST00000541550.1 |
ADK
|
adenosine kinase |
| chr10_+_13203543 | 10.19 |
ENST00000378714.3
ENST00000479669.1 ENST00000484800.2 |
MCM10
|
minichromosome maintenance complex component 10 |
| chr1_+_26798955 | 10.18 |
ENST00000361427.5
|
HMGN2
|
high mobility group nucleosomal binding domain 2 |
| chr6_-_8102714 | 9.97 |
ENST00000502429.1
ENST00000429723.2 ENST00000507463.1 ENST00000379715.5 |
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr4_+_107236722 | 9.66 |
ENST00000442366.1
|
AIMP1
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chr4_+_107236847 | 9.65 |
ENST00000358008.3
|
AIMP1
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chr1_-_51425902 | 9.54 |
ENST00000396153.2
|
FAF1
|
Fas (TNFRSF6) associated factor 1 |
| chr2_+_238395042 | 9.51 |
ENST00000429898.1
ENST00000410032.1 |
MLPH
|
melanophilin |
| chr2_-_152146385 | 9.43 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr5_+_65222384 | 9.34 |
ENST00000380943.2
ENST00000416865.2 ENST00000380939.2 ENST00000380936.1 ENST00000380935.1 |
ERBB2IP
|
erbb2 interacting protein |
| chr19_-_44031341 | 9.14 |
ENST00000600651.1
|
ETHE1
|
ethylmalonic encephalopathy 1 |
| chrX_+_64887512 | 9.08 |
ENST00000360270.5
|
MSN
|
moesin |
| chr1_+_29063271 | 8.90 |
ENST00000373812.3
|
YTHDF2
|
YTH domain family, member 2 |
| chr1_+_29063439 | 8.88 |
ENST00000541996.1
ENST00000496288.1 |
YTHDF2
|
YTH domain family, member 2 |
| chr18_+_12308231 | 8.70 |
ENST00000590103.1
ENST00000591909.1 ENST00000586653.1 ENST00000592683.1 ENST00000590967.1 ENST00000591208.1 ENST00000591463.1 |
TUBB6
|
tubulin, beta 6 class V |
| chr9_-_21995300 | 8.47 |
ENST00000498628.2
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr3_+_37035289 | 8.39 |
ENST00000455445.2
ENST00000441265.1 ENST00000435176.1 ENST00000429117.1 ENST00000536378.1 |
MLH1
|
mutL homolog 1 |
| chr11_-_122931881 | 8.38 |
ENST00000526110.1
ENST00000227378.3 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr6_-_31697255 | 8.32 |
ENST00000436437.1
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr11_+_46403303 | 8.16 |
ENST00000407067.1
ENST00000395565.1 |
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr6_+_3000195 | 8.11 |
ENST00000338130.2
|
NQO2
|
NAD(P)H dehydrogenase, quinone 2 |
| chr7_-_47621736 | 8.07 |
ENST00000311160.9
|
TNS3
|
tensin 3 |
| chrX_+_153607557 | 7.92 |
ENST00000369842.4
ENST00000369835.3 |
EMD
|
emerin |
| chr9_-_21994597 | 7.87 |
ENST00000579755.1
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr6_+_31371337 | 7.81 |
ENST00000449934.2
ENST00000421350.1 |
MICA
|
MHC class I polypeptide-related sequence A |
| chr3_+_107241783 | 7.60 |
ENST00000415149.2
ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr1_-_51425772 | 7.49 |
ENST00000371778.4
|
FAF1
|
Fas (TNFRSF6) associated factor 1 |
| chr12_-_76478446 | 7.30 |
ENST00000393263.3
ENST00000548044.1 ENST00000547704.1 ENST00000431879.3 ENST00000549596.1 ENST00000550934.1 ENST00000551600.1 ENST00000547479.1 ENST00000547773.1 ENST00000544816.1 ENST00000542344.1 ENST00000548273.1 |
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr9_-_21994344 | 7.11 |
ENST00000530628.2
ENST00000361570.3 |
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr18_-_54305658 | 7.05 |
ENST00000586262.1
ENST00000217515.6 |
TXNL1
|
thioredoxin-like 1 |
| chr16_-_20817753 | 6.91 |
ENST00000389345.5
ENST00000300005.3 ENST00000357967.4 ENST00000569729.1 |
ERI2
|
ERI1 exoribonuclease family member 2 |
| chr6_-_74231444 | 6.79 |
ENST00000331523.2
ENST00000356303.2 |
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr11_+_46403194 | 6.73 |
ENST00000395569.4
ENST00000395566.4 |
MDK
|
midkine (neurite growth-promoting factor 2) |
| chrX_-_118925600 | 6.71 |
ENST00000361575.3
|
RPL39
|
ribosomal protein L39 |
| chr6_+_3000218 | 6.64 |
ENST00000380441.1
ENST00000380455.4 ENST00000380454.4 |
NQO2
|
NAD(P)H dehydrogenase, quinone 2 |
| chrX_+_151903207 | 6.55 |
ENST00000370287.3
|
CSAG1
|
chondrosarcoma associated gene 1 |
| chr1_-_246729544 | 6.48 |
ENST00000544618.1
ENST00000366514.4 |
TFB2M
|
transcription factor B2, mitochondrial |
| chr4_-_2935674 | 6.42 |
ENST00000514800.1
|
MFSD10
|
major facilitator superfamily domain containing 10 |
| chr11_+_62649158 | 6.41 |
ENST00000539891.1
ENST00000536981.1 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr6_-_31697563 | 6.40 |
ENST00000375789.2
ENST00000416410.1 |
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr5_+_65222299 | 6.35 |
ENST00000284037.5
|
ERBB2IP
|
erbb2 interacting protein |
| chr7_+_96745902 | 6.32 |
ENST00000432641.2
|
ACN9
|
ACN9 homolog (S. cerevisiae) |
| chrX_+_151883090 | 6.26 |
ENST00000370293.2
ENST00000423993.1 ENST00000447530.1 ENST00000458057.1 ENST00000331220.2 ENST00000422085.1 ENST00000453150.1 ENST00000409560.1 |
MAGEA2B
|
melanoma antigen family A, 2B |
| chr3_+_184033135 | 6.21 |
ENST00000424196.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr8_-_95487331 | 6.18 |
ENST00000336148.5
|
RAD54B
|
RAD54 homolog B (S. cerevisiae) |
| chrX_+_151903253 | 6.11 |
ENST00000452779.2
ENST00000370291.2 |
CSAG1
|
chondrosarcoma associated gene 1 |
| chr11_-_28129656 | 6.07 |
ENST00000263181.6
|
KIF18A
|
kinesin family member 18A |
| chr5_-_37371278 | 6.02 |
ENST00000231498.3
|
NUP155
|
nucleoporin 155kDa |
| chr17_+_33914276 | 5.98 |
ENST00000592545.1
ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chrX_+_129535937 | 5.83 |
ENST00000305536.6
ENST00000370947.1 |
RBMX2
|
RNA binding motif protein, X-linked 2 |
| chr11_-_6633799 | 5.71 |
ENST00000299424.4
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa |
| chr2_+_238395803 | 5.70 |
ENST00000264605.3
|
MLPH
|
melanophilin |
| chr5_-_37371163 | 5.64 |
ENST00000513532.1
|
NUP155
|
nucleoporin 155kDa |
| chrX_-_153714994 | 5.62 |
ENST00000369660.4
|
UBL4A
|
ubiquitin-like 4A |
| chr6_-_31697977 | 5.58 |
ENST00000375787.2
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr11_+_2923423 | 5.55 |
ENST00000312221.5
|
SLC22A18
|
solute carrier family 22, member 18 |
| chr11_+_114270752 | 5.35 |
ENST00000540163.1
|
RBM7
|
RNA binding motif protein 7 |
| chr2_+_189156389 | 5.32 |
ENST00000409843.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr11_+_114271314 | 5.28 |
ENST00000541475.1
|
RBM7
|
RNA binding motif protein 7 |
| chr5_-_148930960 | 5.25 |
ENST00000261798.5
ENST00000377843.2 |
CSNK1A1
|
casein kinase 1, alpha 1 |
| chr17_-_20946338 | 5.25 |
ENST00000261497.4
|
USP22
|
ubiquitin specific peptidase 22 |
| chr15_-_22473353 | 5.23 |
ENST00000557788.2
|
IGHV4OR15-8
|
immunoglobulin heavy variable 4/OR15-8 (non-functional) |
| chr6_+_63921351 | 4.94 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr11_+_114271251 | 4.90 |
ENST00000375490.5
|
RBM7
|
RNA binding motif protein 7 |
| chr16_+_70557685 | 4.89 |
ENST00000302516.5
ENST00000566095.2 ENST00000577085.1 ENST00000567654.1 |
SF3B3
|
splicing factor 3b, subunit 3, 130kDa |
| chr6_+_64281906 | 4.89 |
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr2_-_188378368 | 4.85 |
ENST00000392365.1
ENST00000435414.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr11_+_2923499 | 4.76 |
ENST00000449793.2
|
SLC22A18
|
solute carrier family 22, member 18 |
| chr5_-_133561752 | 4.76 |
ENST00000519718.1
ENST00000481195.1 |
CTD-2410N18.5
PPP2CA
|
S-phase kinase-associated protein 1 protein phosphatase 2, catalytic subunit, alpha isozyme |
| chr17_-_46035187 | 4.69 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr3_-_119396193 | 4.64 |
ENST00000484810.1
ENST00000497116.1 ENST00000261070.2 |
COX17
|
COX17 cytochrome c oxidase copper chaperone |
| chr17_-_40540377 | 4.64 |
ENST00000404395.3
ENST00000389272.3 ENST00000585517.1 ENST00000588065.1 |
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr14_-_106406090 | 4.62 |
ENST00000390593.2
|
IGHV6-1
|
immunoglobulin heavy variable 6-1 |
| chr12_-_57522813 | 4.57 |
ENST00000556155.1
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr5_-_176900610 | 4.54 |
ENST00000477391.2
ENST00000393565.1 ENST00000309007.5 |
DBN1
|
drebrin 1 |
| chr2_-_56150184 | 4.52 |
ENST00000394554.1
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr2_+_238395879 | 4.40 |
ENST00000445024.2
ENST00000338530.4 ENST00000409373.1 |
MLPH
|
melanophilin |
| chr1_-_39325431 | 4.36 |
ENST00000373001.3
|
RRAGC
|
Ras-related GTP binding C |
| chr9_+_71736177 | 4.30 |
ENST00000606364.1
ENST00000453658.2 |
TJP2
|
tight junction protein 2 |
| chr5_-_149669192 | 4.30 |
ENST00000398376.3
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr1_-_151162606 | 4.25 |
ENST00000354473.4
ENST00000368892.4 |
VPS72
|
vacuolar protein sorting 72 homolog (S. cerevisiae) |
| chr10_-_33625154 | 4.25 |
ENST00000265371.4
|
NRP1
|
neuropilin 1 |
| chr16_-_28222797 | 4.21 |
ENST00000569951.1
ENST00000565698.1 |
XPO6
|
exportin 6 |
| chr2_+_90060377 | 4.20 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chrX_-_151922340 | 4.20 |
ENST00000370284.1
ENST00000543232.1 ENST00000393876.1 ENST00000393872.3 |
MAGEA2
|
melanoma antigen family A, 2 |
| chr11_+_2923619 | 4.19 |
ENST00000380574.1
|
SLC22A18
|
solute carrier family 22, member 18 |
| chr6_+_64282447 | 4.07 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr9_-_21995249 | 3.96 |
ENST00000494262.1
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr6_+_37012607 | 3.89 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr17_-_40540586 | 3.82 |
ENST00000264657.5
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr21_+_34638656 | 3.81 |
ENST00000290200.2
|
IL10RB
|
interleukin 10 receptor, beta |
| chr18_+_56338750 | 3.78 |
ENST00000345724.3
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
| chrX_+_105937068 | 3.70 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr14_-_106733624 | 3.69 |
ENST00000390610.2
|
IGHV1-24
|
immunoglobulin heavy variable 1-24 |
| chr2_+_232572361 | 3.62 |
ENST00000409321.1
|
PTMA
|
prothymosin, alpha |
| chr14_-_107095662 | 3.33 |
ENST00000390630.2
|
IGHV4-61
|
immunoglobulin heavy variable 4-61 |
| chr3_-_113897899 | 3.30 |
ENST00000383673.2
ENST00000295881.7 |
DRD3
|
dopamine receptor D3 |
| chr14_-_106878083 | 3.29 |
ENST00000390619.2
|
IGHV4-39
|
immunoglobulin heavy variable 4-39 |
| chr21_+_37757668 | 3.26 |
ENST00000314103.5
|
CHAF1B
|
chromatin assembly factor 1, subunit B (p60) |
| chr5_-_132362226 | 3.23 |
ENST00000509437.1
ENST00000355372.2 ENST00000513541.1 ENST00000509008.1 ENST00000513848.1 ENST00000504170.1 ENST00000324170.3 |
ZCCHC10
|
zinc finger, CCHC domain containing 10 |
| chr8_-_95908902 | 3.15 |
ENST00000520509.1
|
CCNE2
|
cyclin E2 |
| chr18_+_56338618 | 3.09 |
ENST00000348428.3
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
| chr19_+_17378278 | 3.02 |
ENST00000596335.1
ENST00000601436.1 ENST00000595632.1 |
BABAM1
|
BRISC and BRCA1 A complex member 1 |
| chr16_-_28223166 | 2.99 |
ENST00000304658.5
|
XPO6
|
exportin 6 |
| chr17_-_5015129 | 2.99 |
ENST00000575898.1
ENST00000416429.2 |
ZNF232
|
zinc finger protein 232 |
| chr3_+_197476621 | 2.98 |
ENST00000241502.4
|
FYTTD1
|
forty-two-three domain containing 1 |
| chrX_-_48776292 | 2.90 |
ENST00000376509.4
|
PIM2
|
pim-2 oncogene |
| chr22_+_23161491 | 2.89 |
ENST00000390316.2
|
IGLV3-9
|
immunoglobulin lambda variable 3-9 (gene/pseudogene) |
| chr19_+_1071203 | 2.86 |
ENST00000543365.1
|
HMHA1
|
histocompatibility (minor) HA-1 |
| chr14_-_106478603 | 2.84 |
ENST00000390596.2
|
IGHV4-4
|
immunoglobulin heavy variable 4-4 |
| chr19_+_17378159 | 2.73 |
ENST00000598188.1
ENST00000359435.4 ENST00000599474.1 ENST00000599057.1 ENST00000601043.1 ENST00000447614.2 |
BABAM1
|
BRISC and BRCA1 A complex member 1 |
| chr2_+_211421262 | 2.63 |
ENST00000233072.5
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr17_-_7082668 | 2.61 |
ENST00000573083.1
ENST00000574388.1 |
ASGR1
|
asialoglycoprotein receptor 1 |
| chr22_+_25003626 | 2.56 |
ENST00000451366.1
ENST00000406383.2 ENST00000428855.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr12_-_86230315 | 2.45 |
ENST00000361228.3
|
RASSF9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr8_-_28243934 | 2.45 |
ENST00000521185.1
ENST00000520290.1 ENST00000344423.5 |
ZNF395
|
zinc finger protein 395 |
| chr5_-_111092873 | 2.39 |
ENST00000509025.1
ENST00000515855.1 |
NREP
|
neuronal regeneration related protein |
| chr14_+_20811766 | 2.36 |
ENST00000250416.5
ENST00000527915.1 |
PARP2
|
poly (ADP-ribose) polymerase 2 |
| chr14_-_106791536 | 2.30 |
ENST00000390613.2
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr14_+_96722539 | 2.29 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr15_+_58430368 | 2.26 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr17_-_40540484 | 2.26 |
ENST00000588969.1
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr19_-_4338783 | 2.13 |
ENST00000601482.1
ENST00000600324.1 |
STAP2
|
signal transducing adaptor family member 2 |
| chr1_+_86889769 | 2.11 |
ENST00000370565.4
|
CLCA2
|
chloride channel accessory 2 |
| chr14_+_20811722 | 2.03 |
ENST00000429687.3
|
PARP2
|
poly (ADP-ribose) polymerase 2 |
| chr2_-_88427568 | 2.02 |
ENST00000393750.3
ENST00000295834.3 |
FABP1
|
fatty acid binding protein 1, liver |
| chr1_-_36947120 | 1.97 |
ENST00000361632.4
|
CSF3R
|
colony stimulating factor 3 receptor (granulocyte) |
| chr1_-_3447967 | 1.87 |
ENST00000294599.4
|
MEGF6
|
multiple EGF-like-domains 6 |
| chr1_-_21503337 | 1.84 |
ENST00000400422.1
ENST00000602326.1 ENST00000411888.1 ENST00000438975.1 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr2_+_108905325 | 1.79 |
ENST00000438339.1
ENST00000409880.1 ENST00000437390.2 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr19_-_47220335 | 1.77 |
ENST00000601806.1
ENST00000593363.1 ENST00000598633.1 ENST00000595515.1 ENST00000433867.1 |
PRKD2
|
protein kinase D2 |
| chr11_+_46402744 | 1.77 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr3_-_142297668 | 1.75 |
ENST00000350721.4
ENST00000383101.3 |
ATR
|
ataxia telangiectasia and Rad3 related |
| chr11_+_120107344 | 1.69 |
ENST00000260264.4
|
POU2F3
|
POU class 2 homeobox 3 |
| chr15_+_59903975 | 1.66 |
ENST00000560585.1
ENST00000396065.1 |
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr1_-_153513170 | 1.61 |
ENST00000368717.2
|
S100A5
|
S100 calcium binding protein A5 |
| chr5_-_111093081 | 1.58 |
ENST00000453526.2
ENST00000509427.1 |
NREP
|
neuronal regeneration related protein |
| chr18_-_60987220 | 1.53 |
ENST00000398117.1
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr9_-_104198042 | 1.53 |
ENST00000374855.4
|
ALDOB
|
aldolase B, fructose-bisphosphate |
| chr5_-_111093340 | 1.50 |
ENST00000508870.1
|
NREP
|
neuronal regeneration related protein |
| chr17_-_80408569 | 1.45 |
ENST00000577696.1
ENST00000577471.1 ENST00000582545.2 ENST00000437807.2 ENST00000583617.1 ENST00000578913.1 ENST00000336995.7 ENST00000577834.1 ENST00000342572.8 ENST00000585064.1 ENST00000585080.1 ENST00000578919.1 ENST00000306645.5 ENST00000434650.2 |
C17orf62
|
chromosome 17 open reading frame 62 |
| chr15_+_75315896 | 1.45 |
ENST00000342932.3
ENST00000564923.1 ENST00000569562.1 ENST00000568649.1 |
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr3_-_197476560 | 1.44 |
ENST00000273582.5
|
KIAA0226
|
KIAA0226 |
| chr17_-_7082861 | 1.38 |
ENST00000269299.3
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr5_-_111093167 | 1.35 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr17_+_75181292 | 1.35 |
ENST00000431431.2
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae) |
| chr14_+_73603126 | 1.32 |
ENST00000557356.1
ENST00000556864.1 ENST00000556533.1 ENST00000556951.1 ENST00000557293.1 ENST00000553719.1 ENST00000553599.1 ENST00000556011.1 ENST00000394157.3 ENST00000357710.4 ENST00000324501.5 ENST00000560005.2 ENST00000555254.1 ENST00000261970.3 ENST00000344094.3 ENST00000554131.1 ENST00000557037.1 |
PSEN1
|
presenilin 1 |
| chr14_-_106830057 | 1.31 |
ENST00000390616.2
|
IGHV4-34
|
immunoglobulin heavy variable 4-34 |
| chr14_-_82089405 | 1.29 |
ENST00000554211.1
|
RP11-799P8.1
|
RP11-799P8.1 |
| chr1_+_73771844 | 1.28 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
| chr2_+_198318147 | 1.23 |
ENST00000263960.2
|
COQ10B
|
coenzyme Q10 homolog B (S. cerevisiae) |
| chrX_+_149009941 | 1.17 |
ENST00000535454.1
ENST00000542674.1 ENST00000286482.1 |
MAGEA8
|
melanoma antigen family A, 8 |
| chrX_+_129040094 | 1.17 |
ENST00000425117.2
|
UTP14A
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr14_+_22320634 | 1.17 |
ENST00000390435.1
|
TRAV8-3
|
T cell receptor alpha variable 8-3 |
| chr19_-_4338838 | 1.16 |
ENST00000594605.1
|
STAP2
|
signal transducing adaptor family member 2 |
| chr8_+_120428546 | 1.11 |
ENST00000259526.3
|
NOV
|
nephroblastoma overexpressed |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFCP2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.1 | 57.0 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 6.1 | 18.3 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 5.8 | 23.4 | GO:0031081 | nuclear pore distribution(GO:0031081) nuclear pore localization(GO:0051664) |
| 5.8 | 17.4 | GO:0002188 | translation reinitiation(GO:0002188) |
| 3.6 | 10.7 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 3.4 | 10.3 | GO:0044209 | AMP salvage(GO:0044209) |
| 3.3 | 33.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 3.2 | 31.6 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 3.1 | 9.4 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 3.1 | 15.7 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 2.9 | 11.7 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 2.9 | 17.2 | GO:1901029 | adenine transport(GO:0015853) negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 2.8 | 8.4 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 2.8 | 16.7 | GO:0030421 | defecation(GO:0030421) |
| 2.5 | 17.8 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 2.5 | 27.4 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 2.4 | 68.4 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 2.3 | 6.9 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 2.2 | 40.0 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 2.2 | 15.2 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 2.1 | 25.7 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 2.1 | 14.9 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 2.0 | 7.8 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 1.7 | 8.4 | GO:1904764 | late endosomal microautophagy(GO:0061738) negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 1.6 | 11.4 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 1.6 | 6.5 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 1.6 | 14.4 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 1.5 | 12.4 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 1.5 | 9.1 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 1.5 | 9.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 1.5 | 4.4 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 1.4 | 10.0 | GO:0090343 | positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 1.4 | 10.9 | GO:0046618 | drug export(GO:0046618) |
| 1.3 | 19.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 1.3 | 6.4 | GO:0060356 | leucine import(GO:0060356) |
| 1.3 | 20.3 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 1.2 | 6.0 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 1.1 | 4.6 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 1.1 | 18.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 1.1 | 17.0 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 1.1 | 12.7 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.9 | 26.4 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.9 | 5.6 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.9 | 22.9 | GO:0070935 | negative regulation of telomere maintenance via telomerase(GO:0032211) 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.8 | 4.2 | GO:1905040 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.8 | 3.3 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) gastric motility(GO:0035482) gastric emptying(GO:0035483) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.8 | 4.5 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.8 | 10.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.8 | 5.3 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.7 | 36.6 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.7 | 8.3 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.6 | 5.5 | GO:2000507 | cap-dependent translational initiation(GO:0002191) positive regulation of energy homeostasis(GO:2000507) |
| 0.6 | 4.8 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.6 | 1.8 | GO:1904882 | establishment of RNA localization to telomere(GO:0097694) establishment of macromolecular complex localization to telomere(GO:0097695) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.6 | 1.1 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.6 | 13.3 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.6 | 6.1 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.5 | 44.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.5 | 1.5 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.5 | 14.6 | GO:0006295 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) telomere maintenance via semi-conservative replication(GO:0032201) error-free translesion synthesis(GO:0070987) |
| 0.5 | 4.5 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.5 | 2.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) urea transmembrane transport(GO:0071918) purine nucleobase transmembrane transport(GO:1904823) |
| 0.4 | 1.8 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.4 | 4.3 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.4 | 4.3 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.4 | 10.6 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.4 | 3.7 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.4 | 6.7 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.4 | 1.5 | GO:0032848 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.4 | 2.6 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.4 | 6.8 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.4 | 4.6 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.3 | 2.3 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.3 | 3.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 13.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.3 | 5.7 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.3 | 10.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 4.2 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.3 | 11.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.3 | 15.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 5.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.2 | 10.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.2 | 0.7 | GO:0015920 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) lipopolysaccharide transport(GO:0015920) |
| 0.2 | 0.7 | GO:0036484 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.2 | 2.7 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 5.7 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.2 | 14.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 2.0 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 | 4.4 | GO:1903432 | regulation of TORC1 signaling(GO:1903432) |
| 0.2 | 10.2 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.2 | 0.4 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) regulation of respiratory burst involved in inflammatory response(GO:0060264) |
| 0.2 | 1.7 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.2 | 6.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 1.6 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.9 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 | 1.7 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 1.0 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.1 | 0.2 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.1 | 2.9 | GO:0007140 | male meiosis(GO:0007140) |
| 0.1 | 4.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 2.0 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 4.2 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 8.8 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 4.8 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 5.7 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 9.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 6.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.2 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.1 | 1.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.7 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.1 | 1.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 1.0 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 0.5 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.1 | 2.8 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.1 | 2.0 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.1 | 1.1 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 2.3 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.0 | 3.3 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 3.8 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 4.9 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.0 | 1.8 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 4.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.8 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 1.2 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.4 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.4 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 36.6 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 4.3 | 12.8 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 3.1 | 18.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) extrinsic component of endosome membrane(GO:0031313) |
| 2.9 | 11.7 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 2.9 | 17.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 2.8 | 8.4 | GO:0005715 | late recombination nodule(GO:0005715) |
| 2.6 | 15.7 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 2.5 | 27.4 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 2.5 | 29.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 1.9 | 37.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 1.8 | 12.4 | GO:0031415 | NatA complex(GO:0031415) |
| 1.4 | 5.6 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 1.4 | 6.9 | GO:0032449 | CBM complex(GO:0032449) |
| 1.3 | 79.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 1.3 | 7.9 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 1.2 | 17.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 1.2 | 8.4 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 1.1 | 10.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 1.0 | 11.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 1.0 | 14.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 1.0 | 10.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.8 | 3.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.8 | 22.9 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.8 | 3.1 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.7 | 4.4 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.7 | 5.7 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.7 | 6.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.6 | 5.7 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.6 | 10.0 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.6 | 6.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.5 | 5.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.4 | 4.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) sorting endosome(GO:0097443) |
| 0.4 | 7.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.4 | 6.0 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.4 | 5.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.3 | 9.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.3 | 4.9 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.3 | 26.0 | GO:0018995 | host(GO:0018995) host cell(GO:0043657) |
| 0.3 | 5.8 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.3 | 14.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.3 | 17.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.3 | 2.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 25.1 | GO:0005901 | caveola(GO:0005901) |
| 0.2 | 6.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 27.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.2 | 9.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 2.5 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.2 | 17.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.2 | 98.6 | GO:0000785 | chromatin(GO:0000785) |
| 0.2 | 15.2 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.2 | 3.3 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.1 | 15.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 4.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 13.7 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 5.6 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 8.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 2.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 4.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 4.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 3.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.5 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 4.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.8 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 3.7 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 4.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.3 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.9 | 31.6 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 6.6 | 26.4 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 6.1 | 18.4 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 4.3 | 17.2 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 4.1 | 20.3 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 3.8 | 11.4 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 3.3 | 33.4 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 3.1 | 40.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 3.1 | 18.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 2.9 | 14.5 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 2.8 | 30.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 2.6 | 15.7 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 1.9 | 5.7 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 1.8 | 14.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 1.8 | 37.0 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 1.6 | 71.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 1.5 | 27.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 1.5 | 4.4 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 1.4 | 8.4 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 1.4 | 11.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 1.3 | 35.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 1.3 | 6.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 1.1 | 14.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 1.1 | 13.3 | GO:0089720 | caspase binding(GO:0089720) |
| 1.1 | 10.7 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 1.0 | 7.0 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 1.0 | 12.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.9 | 4.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.9 | 2.6 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.9 | 7.8 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.8 | 4.6 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.8 | 2.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.8 | 8.4 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.7 | 34.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.6 | 6.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.6 | 4.2 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.6 | 10.3 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.6 | 10.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.6 | 17.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.6 | 2.3 | GO:0015254 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.6 | 1.7 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.5 | 3.8 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.5 | 6.4 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.5 | 3.6 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.5 | 4.0 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.5 | 2.0 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.5 | 10.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.5 | 3.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.4 | 37.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.4 | 2.3 | GO:0047115 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 0.4 | 5.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.4 | 9.1 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.3 | 1.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.3 | 5.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.3 | 2.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 19.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.3 | 6.0 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.3 | 14.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.3 | 4.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 4.3 | GO:0010857 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.2 | 4.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 1.3 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.2 | 9.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 1.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 0.8 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.2 | 10.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 1.8 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.2 | 7.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 7.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 0.8 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 2.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 4.9 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 1.0 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.2 | 15.5 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.2 | 10.9 | GO:0008186 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.2 | 14.6 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.2 | 10.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 9.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 1.6 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.1 | 2.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 1.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 3.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 12.8 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 5.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 0.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 4.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.7 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.4 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 10.6 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 2.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.8 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 16.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 6.9 | GO:0019209 | kinase activator activity(GO:0019209) |
| 0.1 | 0.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 4.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 5.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 11.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 9.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 4.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 4.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 3.0 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 7.4 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 30.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 1.2 | 30.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 1.1 | 73.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.6 | 10.7 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.4 | 9.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.4 | 21.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 11.4 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.2 | 4.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 6.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.2 | 9.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.2 | 22.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.2 | 19.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.2 | 6.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 6.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 5.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 4.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 12.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 8.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 5.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 0.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 19.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 7.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 8.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 4.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 4.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 5.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 7.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.9 | SIG CHEMOTAXIS | Genes related to chemotaxis |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 79.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 2.0 | 79.2 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 1.6 | 31.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 1.6 | 17.2 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 1.4 | 11.1 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 1.3 | 46.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 1.2 | 14.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.8 | 14.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.7 | 14.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.6 | 10.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.5 | 37.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.5 | 32.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.5 | 11.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.4 | 11.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.4 | 10.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 8.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 5.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.3 | 5.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.3 | 10.2 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.3 | 6.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.3 | 4.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 8.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 6.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 3.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 4.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.2 | 1.8 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 27.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.2 | 5.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 2.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 6.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 22.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 6.9 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 2.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 8.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.3 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 2.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 4.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 3.1 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 3.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 4.9 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.1 | 1.0 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 6.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 3.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.3 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |