Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
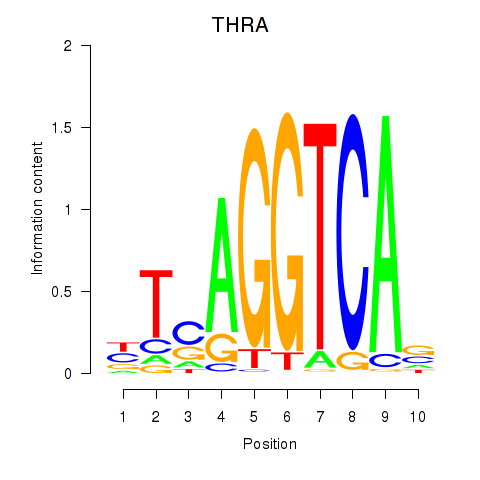
Results for THRA_RXRB
Z-value: 0.96
Transcription factors associated with THRA_RXRB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
THRA
|
ENSG00000126351.8 | thyroid hormone receptor alpha |
|
RXRB
|
ENSG00000204231.6 | retinoid X receptor beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| THRA | hg19_v2_chr17_+_38219063_38219154 | -0.40 | 6.4e-10 | Click! |
| RXRB | hg19_v2_chr6_-_33168391_33168465 | -0.03 | 7.0e-01 | Click! |
Activity profile of THRA_RXRB motif
Sorted Z-values of THRA_RXRB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_23264766 | 47.23 |
ENST00000390331.2
|
IGLC7
|
immunoglobulin lambda constant 7 |
| chr14_-_106054659 | 33.86 |
ENST00000390539.2
|
IGHA2
|
immunoglobulin heavy constant alpha 2 (A2m marker) |
| chr22_+_23165153 | 22.82 |
ENST00000390317.2
|
IGLV2-8
|
immunoglobulin lambda variable 2-8 |
| chr22_+_23134974 | 18.64 |
ENST00000390314.2
|
IGLV2-11
|
immunoglobulin lambda variable 2-11 |
| chr19_-_39826639 | 18.38 |
ENST00000602185.1
ENST00000598034.1 ENST00000601387.1 ENST00000595636.1 ENST00000253054.8 ENST00000594700.1 ENST00000597595.1 |
GMFG
|
glia maturation factor, gamma |
| chr14_-_106174960 | 17.96 |
ENST00000390547.2
|
IGHA1
|
immunoglobulin heavy constant alpha 1 |
| chr22_+_23243156 | 16.40 |
ENST00000390323.2
|
IGLC2
|
immunoglobulin lambda constant 2 (Kern-Oz- marker) |
| chr22_+_23077065 | 15.38 |
ENST00000390310.2
|
IGLV2-18
|
immunoglobulin lambda variable 2-18 |
| chr16_+_85942594 | 14.93 |
ENST00000566369.1
|
IRF8
|
interferon regulatory factor 8 |
| chr19_-_10450328 | 14.65 |
ENST00000160262.5
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr19_-_10450287 | 12.75 |
ENST00000589261.1
ENST00000590569.1 ENST00000589580.1 ENST00000589249.1 |
ICAM3
|
intercellular adhesion molecule 3 |
| chr2_-_230786679 | 12.18 |
ENST00000543084.1
ENST00000343290.5 ENST00000389044.4 ENST00000283943.5 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr22_+_22681656 | 11.83 |
ENST00000390291.2
|
IGLV1-50
|
immunoglobulin lambda variable 1-50 (non-functional) |
| chr22_+_22712087 | 11.38 |
ENST00000390294.2
|
IGLV1-47
|
immunoglobulin lambda variable 1-47 |
| chr6_+_139456226 | 10.76 |
ENST00000367658.2
|
HECA
|
headcase homolog (Drosophila) |
| chr22_+_23040274 | 10.64 |
ENST00000390306.2
|
IGLV2-23
|
immunoglobulin lambda variable 2-23 |
| chr22_+_22764088 | 9.41 |
ENST00000390299.2
|
IGLV1-40
|
immunoglobulin lambda variable 1-40 |
| chr2_-_230786619 | 9.09 |
ENST00000389045.3
ENST00000409677.1 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr1_-_149900122 | 8.87 |
ENST00000271628.8
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa |
| chr19_+_1065922 | 8.21 |
ENST00000539243.2
|
HMHA1
|
histocompatibility (minor) HA-1 |
| chrX_-_152989798 | 7.79 |
ENST00000441714.1
ENST00000442093.1 ENST00000429550.1 ENST00000345046.6 |
BCAP31
|
B-cell receptor-associated protein 31 |
| chr14_-_55369525 | 7.61 |
ENST00000543643.2
ENST00000536224.2 ENST00000395514.1 ENST00000491895.2 |
GCH1
|
GTP cyclohydrolase 1 |
| chr2_+_87565634 | 7.30 |
ENST00000421835.2
|
IGKV3OR2-268
|
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
| chr19_+_49838653 | 7.15 |
ENST00000598095.1
ENST00000426897.2 ENST00000323906.4 ENST00000535669.2 ENST00000597602.1 ENST00000595660.1 |
CD37
|
CD37 molecule |
| chr13_-_41635512 | 7.14 |
ENST00000405737.2
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr2_-_90538397 | 7.10 |
ENST00000443397.3
|
RP11-685N3.1
|
Uncharacterized protein |
| chr2_+_90153696 | 7.06 |
ENST00000417279.2
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 (gene/pseudogene) |
| chr5_-_149829314 | 6.93 |
ENST00000407193.1
|
RPS14
|
ribosomal protein S14 |
| chr2_-_176046391 | 6.81 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr5_-_149829294 | 6.70 |
ENST00000401695.3
|
RPS14
|
ribosomal protein S14 |
| chr13_+_76123883 | 6.07 |
ENST00000377595.3
|
UCHL3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr16_+_85646763 | 5.99 |
ENST00000411612.1
ENST00000253458.7 |
GSE1
|
Gse1 coiled-coil protein |
| chr13_-_46756351 | 5.98 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr1_+_41204506 | 5.95 |
ENST00000525290.1
ENST00000530965.1 ENST00000416859.2 ENST00000308733.5 |
NFYC
|
nuclear transcription factor Y, gamma |
| chr22_-_36784035 | 5.92 |
ENST00000216181.5
|
MYH9
|
myosin, heavy chain 9, non-muscle |
| chr11_-_58343319 | 5.36 |
ENST00000395074.2
|
LPXN
|
leupaxin |
| chr12_-_102513843 | 5.31 |
ENST00000551744.2
ENST00000552283.1 |
NUP37
|
nucleoporin 37kDa |
| chr14_-_104028595 | 5.16 |
ENST00000337322.4
ENST00000445922.2 |
BAG5
|
BCL2-associated athanogene 5 |
| chr5_-_180666570 | 5.10 |
ENST00000509535.1
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr20_+_43343886 | 5.05 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr14_-_58893832 | 5.04 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr14_+_22963806 | 4.93 |
ENST00000390493.1
|
TRAJ44
|
T cell receptor alpha joining 44 |
| chr3_-_127872625 | 4.84 |
ENST00000464873.1
|
RUVBL1
|
RuvB-like AAA ATPase 1 |
| chr13_-_95131923 | 4.65 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr17_-_18161870 | 4.63 |
ENST00000579294.1
ENST00000545457.2 ENST00000379450.4 ENST00000578558.1 |
FLII
|
flightless I homolog (Drosophila) |
| chr22_+_22676808 | 4.60 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr5_-_149829244 | 4.55 |
ENST00000312037.5
|
RPS14
|
ribosomal protein S14 |
| chrX_-_129299638 | 4.05 |
ENST00000535724.1
ENST00000346424.2 |
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1 |
| chr9_-_139305051 | 3.96 |
ENST00000371725.3
ENST00000298537.7 |
SDCCAG3
|
serologically defined colon cancer antigen 3 |
| chr17_+_8191815 | 3.72 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr3_-_195270162 | 3.61 |
ENST00000438848.1
ENST00000328432.3 |
PPP1R2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr19_+_6739662 | 3.58 |
ENST00000313285.8
ENST00000313244.9 ENST00000596758.1 |
TRIP10
|
thyroid hormone receptor interactor 10 |
| chr20_+_3776371 | 3.58 |
ENST00000245960.5
|
CDC25B
|
cell division cycle 25B |
| chr14_-_104029013 | 3.56 |
ENST00000299204.4
ENST00000557666.1 |
BAG5
|
BCL2-associated athanogene 5 |
| chr6_+_21593972 | 3.52 |
ENST00000244745.1
ENST00000543472.1 |
SOX4
|
SRY (sex determining region Y)-box 4 |
| chr22_-_21581926 | 3.44 |
ENST00000401924.1
|
GGT2
|
gamma-glutamyltransferase 2 |
| chr14_-_24616426 | 3.37 |
ENST00000216802.5
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr12_+_56211703 | 3.33 |
ENST00000243045.5
ENST00000552672.1 ENST00000550836.1 |
ORMDL2
|
ORM1-like 2 (S. cerevisiae) |
| chr1_+_155583012 | 3.30 |
ENST00000462250.2
|
MSTO1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr20_+_3776936 | 3.24 |
ENST00000439880.2
|
CDC25B
|
cell division cycle 25B |
| chr7_+_5085452 | 3.23 |
ENST00000353796.3
ENST00000396912.1 ENST00000396904.2 |
RBAK
RBAK-RBAKDN
|
RB-associated KRAB zinc finger RBAK-RBAKDN readthrough |
| chr2_-_179315786 | 3.07 |
ENST00000457633.1
ENST00000438687.3 ENST00000325748.4 |
PRKRA
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr20_+_35807512 | 2.98 |
ENST00000373622.5
|
RPN2
|
ribophorin II |
| chr9_+_116298778 | 2.94 |
ENST00000462143.1
|
RGS3
|
regulator of G-protein signaling 3 |
| chr2_-_179315490 | 2.92 |
ENST00000487082.1
|
PRKRA
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr20_+_35807449 | 2.92 |
ENST00000237530.6
|
RPN2
|
ribophorin II |
| chr22_-_29137771 | 2.91 |
ENST00000439200.1
ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2
|
checkpoint kinase 2 |
| chr16_+_3074002 | 2.80 |
ENST00000326266.8
ENST00000574549.1 ENST00000575576.1 ENST00000253952.9 |
THOC6
|
THO complex 6 homolog (Drosophila) |
| chr1_+_45212074 | 2.76 |
ENST00000372217.1
|
KIF2C
|
kinesin family member 2C |
| chr17_+_27071002 | 2.73 |
ENST00000262395.5
ENST00000422344.1 ENST00000444415.3 ENST00000262396.6 |
TRAF4
|
TNF receptor-associated factor 4 |
| chr2_+_55459495 | 2.70 |
ENST00000272317.6
ENST00000449323.1 |
RPS27A
|
ribosomal protein S27a |
| chr9_+_36572851 | 2.64 |
ENST00000298048.2
ENST00000538311.1 ENST00000536987.1 ENST00000545008.1 ENST00000536860.1 ENST00000536329.1 ENST00000541717.1 ENST00000543751.1 |
MELK
|
maternal embryonic leucine zipper kinase |
| chrX_+_9431324 | 2.62 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr2_-_179315453 | 2.57 |
ENST00000432031.2
|
PRKRA
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr19_+_58898627 | 2.56 |
ENST00000598098.1
ENST00000598495.1 ENST00000196551.3 ENST00000596046.1 |
RPS5
|
ribosomal protein S5 |
| chr14_-_24036943 | 2.55 |
ENST00000556843.1
ENST00000397120.3 ENST00000557189.1 |
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit |
| chr9_+_131452239 | 2.54 |
ENST00000372688.4
ENST00000372686.5 |
SET
|
SET nuclear oncogene |
| chr1_+_45212051 | 2.52 |
ENST00000372222.3
|
KIF2C
|
kinesin family member 2C |
| chr2_-_183387283 | 2.51 |
ENST00000435564.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr16_+_85646891 | 2.48 |
ENST00000393243.1
|
GSE1
|
Gse1 coiled-coil protein |
| chr18_+_19749386 | 2.48 |
ENST00000269216.3
|
GATA6
|
GATA binding protein 6 |
| chr8_+_103563792 | 2.47 |
ENST00000285402.3
|
ODF1
|
outer dense fiber of sperm tails 1 |
| chr8_+_110552831 | 2.47 |
ENST00000530629.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr11_-_5531215 | 2.43 |
ENST00000311659.4
|
UBQLN3
|
ubiquilin 3 |
| chr1_+_203274639 | 2.40 |
ENST00000290551.4
|
BTG2
|
BTG family, member 2 |
| chr18_+_3262954 | 2.38 |
ENST00000584539.1
|
MYL12B
|
myosin, light chain 12B, regulatory |
| chr1_-_175162048 | 2.38 |
ENST00000444639.1
|
KIAA0040
|
KIAA0040 |
| chr7_+_73245193 | 2.33 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr17_-_41132010 | 2.32 |
ENST00000409103.1
ENST00000360221.4 |
PTGES3L-AARSD1
|
PTGES3L-AARSD1 readthrough |
| chr9_+_117350009 | 2.31 |
ENST00000374050.3
|
ATP6V1G1
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1 |
| chr9_+_139305102 | 2.29 |
ENST00000371720.1
ENST00000371717.3 ENST00000399219.3 |
PMPCA
|
peptidase (mitochondrial processing) alpha |
| chr1_-_151431647 | 2.23 |
ENST00000368863.2
ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ
|
pogo transposable element with ZNF domain |
| chr9_+_34652164 | 2.19 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr17_-_5389477 | 2.17 |
ENST00000572834.1
ENST00000570848.1 ENST00000571971.1 ENST00000158771.4 |
DERL2
|
derlin 2 |
| chr15_-_75230368 | 2.17 |
ENST00000564811.1
ENST00000562233.1 ENST00000567270.1 ENST00000568783.1 |
COX5A
|
cytochrome c oxidase subunit Va |
| chr7_-_140179276 | 2.16 |
ENST00000443720.2
ENST00000255977.2 |
MKRN1
|
makorin ring finger protein 1 |
| chr3_+_184038073 | 2.09 |
ENST00000428387.1
ENST00000434061.2 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr1_+_93544821 | 2.09 |
ENST00000370303.4
|
MTF2
|
metal response element binding transcription factor 2 |
| chr1_-_33815486 | 2.07 |
ENST00000373418.3
|
PHC2
|
polyhomeotic homolog 2 (Drosophila) |
| chr15_+_44084503 | 2.03 |
ENST00000409960.2
ENST00000409646.1 ENST00000594896.1 ENST00000339624.5 ENST00000409291.1 ENST00000402131.1 ENST00000403425.1 ENST00000430901.1 |
SERF2
|
small EDRK-rich factor 2 |
| chr3_+_25831567 | 1.96 |
ENST00000280701.3
ENST00000420173.2 |
OXSM
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr10_+_81107271 | 1.91 |
ENST00000448165.1
|
PPIF
|
peptidylprolyl isomerase F |
| chr10_+_11207438 | 1.90 |
ENST00000609692.1
ENST00000354897.3 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr11_-_59578202 | 1.88 |
ENST00000300151.4
|
MRPL16
|
mitochondrial ribosomal protein L16 |
| chr2_+_86426478 | 1.87 |
ENST00000254644.8
ENST00000605125.1 ENST00000337109.4 ENST00000409180.1 |
MRPL35
|
mitochondrial ribosomal protein L35 |
| chr11_+_117049854 | 1.86 |
ENST00000278951.7
|
SIDT2
|
SID1 transmembrane family, member 2 |
| chr15_+_44084040 | 1.82 |
ENST00000249786.4
|
SERF2
|
small EDRK-rich factor 2 |
| chr5_+_139927213 | 1.79 |
ENST00000310331.2
|
EIF4EBP3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr9_-_139304979 | 1.79 |
ENST00000357365.3
ENST00000371723.4 |
SDCCAG3
|
serologically defined colon cancer antigen 3 |
| chr1_-_103574024 | 1.79 |
ENST00000512756.1
ENST00000370096.3 ENST00000358392.2 ENST00000353414.4 |
COL11A1
|
collagen, type XI, alpha 1 |
| chr2_-_183387430 | 1.77 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr22_+_30163340 | 1.76 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr6_+_43457317 | 1.75 |
ENST00000438588.2
|
TJAP1
|
tight junction associated protein 1 (peripheral) |
| chr2_-_100106419 | 1.73 |
ENST00000393445.3
ENST00000258428.3 |
REV1
|
REV1, polymerase (DNA directed) |
| chr11_-_64013663 | 1.72 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr19_-_40228657 | 1.68 |
ENST00000221804.4
|
CLC
|
Charcot-Leyden crystal galectin |
| chr11_+_122709200 | 1.67 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr2_-_192016316 | 1.63 |
ENST00000358470.4
ENST00000432798.1 ENST00000450994.1 |
STAT4
|
signal transducer and activator of transcription 4 |
| chr17_-_41132410 | 1.60 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr3_+_184037466 | 1.60 |
ENST00000441154.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr11_+_71935797 | 1.59 |
ENST00000298229.2
ENST00000541756.1 |
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr19_+_16186903 | 1.58 |
ENST00000588507.1
|
TPM4
|
tropomyosin 4 |
| chr19_+_39989535 | 1.57 |
ENST00000356433.5
|
DLL3
|
delta-like 3 (Drosophila) |
| chr11_+_7618413 | 1.57 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr2_-_183387064 | 1.56 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr19_-_52227221 | 1.55 |
ENST00000222115.1
ENST00000540069.2 |
HAS1
|
hyaluronan synthase 1 |
| chr22_-_29138386 | 1.54 |
ENST00000544772.1
|
CHEK2
|
checkpoint kinase 2 |
| chr10_+_75757863 | 1.52 |
ENST00000372755.3
ENST00000211998.4 ENST00000417648.2 |
VCL
|
vinculin |
| chr10_+_104178946 | 1.50 |
ENST00000432590.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr10_+_11784360 | 1.47 |
ENST00000379215.4
ENST00000420401.1 |
ECHDC3
|
enoyl CoA hydratase domain containing 3 |
| chr1_+_55107449 | 1.44 |
ENST00000421030.2
ENST00000545244.1 ENST00000339553.5 ENST00000409996.1 ENST00000454855.2 |
MROH7
|
maestro heat-like repeat family member 7 |
| chr12_-_114843889 | 1.37 |
ENST00000405440.2
|
TBX5
|
T-box 5 |
| chr14_+_81421921 | 1.37 |
ENST00000554263.1
ENST00000554435.1 |
TSHR
|
thyroid stimulating hormone receptor |
| chr4_-_151936416 | 1.36 |
ENST00000510413.1
ENST00000507224.1 |
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr12_+_52626898 | 1.30 |
ENST00000331817.5
|
KRT7
|
keratin 7 |
| chr2_+_45168875 | 1.30 |
ENST00000260653.3
|
SIX3
|
SIX homeobox 3 |
| chr2_+_201936707 | 1.30 |
ENST00000433898.1
ENST00000454214.1 |
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr14_+_81421861 | 1.30 |
ENST00000298171.2
|
TSHR
|
thyroid stimulating hormone receptor |
| chr16_-_47007545 | 1.29 |
ENST00000317089.5
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2 |
| chrX_-_39956656 | 1.28 |
ENST00000397354.3
ENST00000378444.4 |
BCOR
|
BCL6 corepressor |
| chr15_-_34629922 | 1.28 |
ENST00000559484.1
ENST00000354181.3 ENST00000558589.1 ENST00000458406.2 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr7_+_100187196 | 1.28 |
ENST00000468962.1
ENST00000427939.2 |
FBXO24
|
F-box protein 24 |
| chr15_-_75017711 | 1.27 |
ENST00000567032.1
ENST00000564596.1 ENST00000566503.1 ENST00000395049.4 ENST00000395048.2 ENST00000379727.3 |
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1 |
| chr14_+_22964877 | 1.24 |
ENST00000390494.1
|
TRAJ43
|
T cell receptor alpha joining 43 |
| chr17_-_80231557 | 1.19 |
ENST00000392334.2
ENST00000314028.6 |
CSNK1D
|
casein kinase 1, delta |
| chr17_+_34171081 | 1.18 |
ENST00000585577.1
|
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr22_-_24316648 | 1.17 |
ENST00000403754.3
ENST00000430101.2 ENST00000398344.4 |
DDT
|
D-dopachrome tautomerase |
| chr1_-_155880672 | 1.14 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr22_+_38245414 | 1.13 |
ENST00000381683.6
ENST00000414316.1 ENST00000406934.1 ENST00000451427.1 |
EIF3L
|
eukaryotic translation initiation factor 3, subunit L |
| chr3_-_187009798 | 1.13 |
ENST00000337774.5
|
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
| chr3_+_179322481 | 1.12 |
ENST00000259037.3
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr6_-_99797522 | 1.10 |
ENST00000389677.5
|
FAXC
|
failed axon connections homolog (Drosophila) |
| chr14_+_81421710 | 1.07 |
ENST00000342443.6
|
TSHR
|
thyroid stimulating hormone receptor |
| chr2_+_55459808 | 1.05 |
ENST00000404735.1
|
RPS27A
|
ribosomal protein S27a |
| chr9_+_127631399 | 1.05 |
ENST00000259477.6
|
ARPC5L
|
actin related protein 2/3 complex, subunit 5-like |
| chr15_-_31393910 | 0.99 |
ENST00000397795.2
ENST00000256552.6 ENST00000559179.1 |
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr19_+_41117770 | 0.96 |
ENST00000601032.1
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr3_+_49058444 | 0.96 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr11_+_111896320 | 0.95 |
ENST00000531306.1
ENST00000537636.1 |
DLAT
|
dihydrolipoamide S-acetyltransferase |
| chr1_-_85040090 | 0.94 |
ENST00000370630.5
|
CTBS
|
chitobiase, di-N-acetyl- |
| chr17_-_4843206 | 0.94 |
ENST00000576951.1
|
SLC25A11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr3_-_15374659 | 0.93 |
ENST00000426925.1
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr1_+_236558694 | 0.91 |
ENST00000359362.5
|
EDARADD
|
EDAR-associated death domain |
| chr1_+_93544791 | 0.87 |
ENST00000545708.1
ENST00000540243.1 ENST00000370298.4 |
MTF2
|
metal response element binding transcription factor 2 |
| chr11_-_30607819 | 0.87 |
ENST00000448418.2
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chr11_+_65190245 | 0.87 |
ENST00000499732.1
ENST00000501122.2 ENST00000601801.1 |
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr19_+_49866331 | 0.87 |
ENST00000597873.1
|
DKKL1
|
dickkopf-like 1 |
| chr1_-_174992544 | 0.86 |
ENST00000476371.1
|
MRPS14
|
mitochondrial ribosomal protein S14 |
| chr11_-_62752162 | 0.86 |
ENST00000458333.2
ENST00000421062.2 |
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr3_+_179322573 | 0.84 |
ENST00000493866.1
ENST00000472629.1 ENST00000482604.1 |
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr14_-_25078864 | 0.81 |
ENST00000216338.4
ENST00000557220.2 ENST00000382548.4 |
GZMH
|
granzyme H (cathepsin G-like 2, protein h-CCPX) |
| chr10_-_76859247 | 0.80 |
ENST00000472493.2
ENST00000605915.1 ENST00000478873.2 |
DUSP13
|
dual specificity phosphatase 13 |
| chr1_+_22962948 | 0.80 |
ENST00000374642.3
|
C1QA
|
complement component 1, q subcomponent, A chain |
| chr2_-_68479614 | 0.79 |
ENST00000234310.3
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha |
| chr1_-_151431909 | 0.78 |
ENST00000361398.3
ENST00000271715.2 |
POGZ
|
pogo transposable element with ZNF domain |
| chr17_+_37894179 | 0.76 |
ENST00000577695.1
ENST00000309156.4 ENST00000309185.3 |
GRB7
|
growth factor receptor-bound protein 7 |
| chr19_-_45826125 | 0.75 |
ENST00000221476.3
|
CKM
|
creatine kinase, muscle |
| chr9_+_125133315 | 0.73 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr1_-_175161890 | 0.73 |
ENST00000545251.2
ENST00000423313.1 |
KIAA0040
|
KIAA0040 |
| chr11_+_66624527 | 0.72 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr11_-_119599794 | 0.70 |
ENST00000264025.3
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chr7_-_73153122 | 0.70 |
ENST00000458339.1
|
ABHD11
|
abhydrolase domain containing 11 |
| chr18_+_12948000 | 0.70 |
ENST00000585730.1
ENST00000399892.2 ENST00000589446.1 ENST00000587761.1 |
SEH1L
|
SEH1-like (S. cerevisiae) |
| chr5_+_162887556 | 0.69 |
ENST00000393915.4
ENST00000432118.2 ENST00000358715.3 |
HMMR
|
hyaluronan-mediated motility receptor (RHAMM) |
| chr19_+_16187085 | 0.68 |
ENST00000300933.4
|
TPM4
|
tropomyosin 4 |
| chr19_+_39989580 | 0.67 |
ENST00000596614.1
ENST00000205143.4 |
DLL3
|
delta-like 3 (Drosophila) |
| chr2_-_49381646 | 0.67 |
ENST00000346173.3
ENST00000406846.2 |
FSHR
|
follicle stimulating hormone receptor |
| chr7_-_150777949 | 0.67 |
ENST00000482571.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr2_-_49381572 | 0.66 |
ENST00000454032.1
ENST00000304421.4 |
FSHR
|
follicle stimulating hormone receptor |
| chr3_-_38691119 | 0.63 |
ENST00000333535.4
ENST00000413689.1 ENST00000443581.1 ENST00000425664.1 ENST00000451551.2 |
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit |
| chr6_+_30131318 | 0.63 |
ENST00000376688.1
|
TRIM15
|
tripartite motif containing 15 |
| chrX_+_70521584 | 0.62 |
ENST00000373829.3
ENST00000538820.1 |
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2 |
| chr5_-_176981417 | 0.62 |
ENST00000514747.1
ENST00000443375.2 ENST00000329540.5 |
FAM193B
|
family with sequence similarity 193, member B |
| chr3_-_197024965 | 0.62 |
ENST00000392382.2
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr3_-_46000064 | 0.61 |
ENST00000433878.1
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr22_+_41075277 | 0.61 |
ENST00000381433.2
|
MCHR1
|
melanin-concentrating hormone receptor 1 |
| chr2_-_165698521 | 0.61 |
ENST00000409184.3
ENST00000392717.2 ENST00000456693.1 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr11_+_64879317 | 0.59 |
ENST00000526809.1
ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2
|
transmembrane 7 superfamily member 2 |
| chr5_+_112849373 | 0.58 |
ENST00000161863.4
ENST00000515883.1 |
YTHDC2
|
YTH domain containing 2 |
| chr1_-_17380630 | 0.57 |
ENST00000375499.3
|
SDHB
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
Network of associatons between targets according to the STRING database.
First level regulatory network of THRA_RXRB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 51.8 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 3.0 | 21.3 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 2.6 | 18.4 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 2.5 | 7.6 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 2.0 | 5.9 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 1.6 | 7.8 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 1.5 | 4.5 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 1.5 | 8.7 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 1.1 | 3.3 | GO:0090155 | negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.9 | 20.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.9 | 3.7 | GO:1905229 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.9 | 3.7 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.9 | 3.5 | GO:0035910 | N-terminal peptidyl-lysine acetylation(GO:0018076) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.9 | 122.8 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.8 | 2.5 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.7 | 14.9 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.7 | 6.8 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.7 | 5.4 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.7 | 4.7 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.7 | 8.6 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.6 | 1.9 | GO:2000276 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.6 | 4.1 | GO:1904044 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) response to aldosterone(GO:1904044) |
| 0.6 | 2.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.6 | 5.0 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.5 | 5.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.5 | 3.0 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.5 | 1.4 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.4 | 1.3 | GO:0021798 | neuroblast differentiation(GO:0014016) forebrain dorsal/ventral pattern formation(GO:0021798) regulation of neural retina development(GO:0061074) regulation of retina development in camera-type eye(GO:1902866) |
| 0.4 | 1.3 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.4 | 1.3 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) cellular alkene metabolic process(GO:0043449) |
| 0.4 | 8.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.4 | 3.7 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.4 | 1.6 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.4 | 1.2 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.4 | 2.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.4 | 4.8 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.3 | 5.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.3 | 1.3 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.3 | 1.9 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.3 | 2.8 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.3 | 2.2 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.3 | 6.0 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.3 | 1.8 | GO:0035989 | tendon development(GO:0035989) |
| 0.3 | 7.1 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.3 | 2.7 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.3 | 4.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.3 | 5.8 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.2 | 41.5 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.2 | 3.4 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.2 | 1.7 | GO:0002420 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.2 | 3.0 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 1.8 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 | 6.8 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.2 | 1.7 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.2 | 3.3 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.2 | 5.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 0.4 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.2 | 2.0 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.2 | 29.3 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.2 | 1.9 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.2 | 0.6 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.2 | 2.4 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 0.9 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 0.4 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.1 | 1.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.1 | 6.0 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 1.5 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.1 | 0.9 | GO:0008154 | actin polymerization or depolymerization(GO:0008154) actin filament polymerization(GO:0030041) |
| 0.1 | 0.3 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.1 | 0.6 | GO:1901098 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 6.5 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.1 | 2.5 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 5.5 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.3 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 4.0 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 0.3 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.9 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 1.5 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 2.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 1.8 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 1.5 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.1 | 0.3 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 4.9 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.1 | 0.8 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.5 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 1.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 1.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.6 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 1.2 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 1.0 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 1.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 1.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.6 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 10.7 | GO:0030323 | respiratory tube development(GO:0030323) |
| 0.0 | 2.6 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.0 | 3.2 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 13.9 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.9 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 1.2 | GO:0006582 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.7 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 1.0 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0016050 | vesicle organization(GO:0016050) |
| 0.0 | 0.2 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 0.3 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 1.0 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 2.1 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 1.3 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.5 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.4 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 2.1 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.8 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.5 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.9 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.4 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.2 | GO:0051005 | nerve growth factor signaling pathway(GO:0038180) plasma membrane to endosome transport(GO:0048227) negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.2 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 1.8 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.3 | 51.8 | GO:0071746 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 2.0 | 5.9 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.3 | 7.8 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 1.3 | 5.0 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 1.0 | 47.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.8 | 3.3 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.6 | 8.6 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.6 | 3.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.5 | 6.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.5 | 5.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.5 | 4.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.5 | 9.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.4 | 1.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.4 | 6.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.4 | 28.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.4 | 6.0 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.4 | 2.5 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.3 | 8.9 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.3 | 2.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.3 | 2.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.3 | 1.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.3 | 5.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.3 | 4.7 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 1.0 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.2 | 3.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 1.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 8.2 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 1.8 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 0.8 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.2 | 4.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.8 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.6 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 1.9 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.1 | 21.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 3.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 1.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 6.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 3.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.3 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.5 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 3.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 15.8 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.1 | 2.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 2.4 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 1.6 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 1.0 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 3.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 4.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 4.4 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 7.6 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 18.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 5.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.9 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 5.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.5 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 1.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 4.5 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.3 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.2 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 2.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 2.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.0 | GO:0098803 | respiratory chain complex(GO:0098803) |
| 0.0 | 2.7 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.9 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 52.4 | GO:0005576 | extracellular region(GO:0005576) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 18.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 2.3 | 18.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 1.8 | 99.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 1.3 | 7.6 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 1.0 | 5.8 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 1.0 | 8.6 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.8 | 5.8 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.8 | 4.8 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.7 | 8.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.6 | 114.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.6 | 6.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.6 | 5.9 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.6 | 1.8 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.5 | 3.7 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.5 | 3.7 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.5 | 5.3 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.5 | 3.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.5 | 3.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.4 | 19.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.4 | 1.1 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.4 | 3.0 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.4 | 5.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.3 | 1.3 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.3 | 1.9 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.3 | 5.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 6.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.3 | 7.8 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.3 | 1.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 3.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 0.6 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 0.6 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.2 | 0.6 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.2 | 2.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 1.9 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.2 | 0.6 | GO:0051538 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.2 | 0.9 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.2 | 0.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.2 | 33.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 2.7 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 2.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 1.6 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 2.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.6 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 18.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 2.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.3 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 4.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 5.4 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 0.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.9 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.9 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.1 | 0.7 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 3.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 7.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 1.7 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 0.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 10.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 3.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 4.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.9 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 2.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 2.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 5.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 2.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 1.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.6 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 4.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 27.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.5 | 6.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 14.9 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.2 | 7.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 12.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 5.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 3.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 17.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 4.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 4.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 5.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.6 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 2.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 2.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 2.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 4.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 6.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 2.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 2.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 3.7 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.3 | 5.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.3 | 29.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.3 | 21.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.3 | 6.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 5.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 5.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 3.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 8.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 14.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.2 | 6.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.2 | 8.9 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 5.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 4.8 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 3.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 26.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 3.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 1.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 5.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 2.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 6.5 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 1.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 1.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 3.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 2.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |