Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
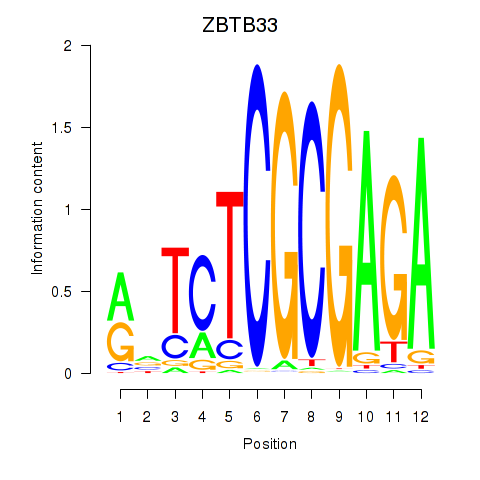
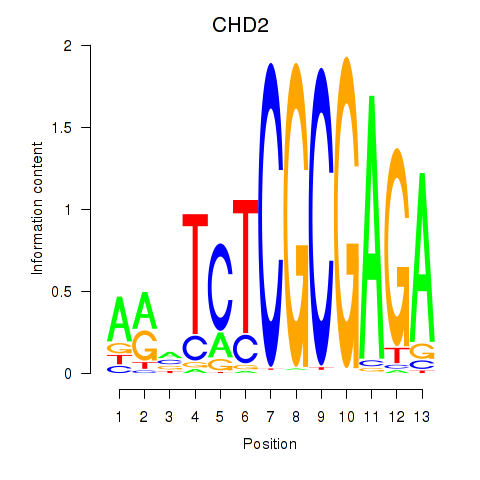
Results for ZBTB33_CHD2
Z-value: 4.62
Transcription factors associated with ZBTB33_CHD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB33
|
ENSG00000177485.6 | zinc finger and BTB domain containing 33 |
|
CHD2
|
ENSG00000173575.14 | chromodomain helicase DNA binding protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CHD2 | hg19_v2_chr15_+_93443419_93443580 | -0.82 | 1.4e-53 | Click! |
| ZBTB33 | hg19_v2_chrX_+_119384607_119384720 | -0.71 | 1.1e-34 | Click! |
Activity profile of ZBTB33_CHD2 motif
Sorted Z-values of ZBTB33_CHD2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB33_CHD2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 43.1 | 129.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 36.7 | 183.6 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 30.9 | 154.7 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 29.1 | 87.4 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 28.7 | 86.2 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 28.0 | 112.1 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 26.6 | 106.5 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 26.3 | 78.9 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 25.0 | 150.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 25.0 | 124.8 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 24.8 | 74.5 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 24.2 | 72.6 | GO:1901189 | arterial endothelial cell fate commitment(GO:0060844) blood vessel lumenization(GO:0072554) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 23.9 | 191.3 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 21.6 | 86.5 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 19.4 | 58.3 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 18.9 | 94.3 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 18.8 | 56.4 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 18.5 | 55.5 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 18.2 | 72.7 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 18.2 | 72.6 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 17.3 | 138.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 17.1 | 102.7 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 16.9 | 169.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 16.2 | 113.6 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 16.0 | 48.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 15.9 | 95.6 | GO:0015853 | adenine transport(GO:0015853) |
| 15.9 | 47.6 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 15.8 | 63.3 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 15.0 | 75.0 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 15.0 | 689.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 14.8 | 89.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 14.2 | 455.0 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 14.0 | 42.1 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 13.9 | 41.7 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 13.7 | 68.7 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 13.5 | 40.4 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 13.3 | 213.2 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 13.0 | 13.0 | GO:0061317 | canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 12.8 | 38.4 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 12.8 | 63.9 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 12.1 | 48.4 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 11.7 | 46.7 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 11.6 | 11.6 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 11.3 | 33.9 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 10.7 | 85.8 | GO:0060699 | regulation of endoribonuclease activity(GO:0060699) |
| 10.7 | 32.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 10.7 | 64.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 10.2 | 50.8 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 9.8 | 127.9 | GO:1990173 | protein localization to nuclear body(GO:1903405) positive regulation of establishment of protein localization to telomere(GO:1904851) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 9.7 | 58.4 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 9.2 | 27.5 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 9.1 | 145.7 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 9.0 | 98.9 | GO:0006983 | ER overload response(GO:0006983) |
| 8.9 | 44.4 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 8.8 | 237.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 8.6 | 25.7 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 8.5 | 110.6 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 8.4 | 83.6 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 8.1 | 8.1 | GO:0061738 | late endosomal microautophagy(GO:0061738) negative regulation of exosomal secretion(GO:1903542) |
| 8.0 | 23.9 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 7.9 | 23.8 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 7.7 | 23.2 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 7.7 | 46.2 | GO:1904044 | response to aldosterone(GO:1904044) |
| 7.6 | 38.2 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 7.6 | 22.9 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 7.6 | 30.4 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 7.1 | 57.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 7.1 | 7.1 | GO:0070203 | regulation of establishment of protein localization to telomere(GO:0070203) |
| 7.0 | 63.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 6.8 | 20.4 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 6.7 | 26.8 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 6.6 | 19.9 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 6.5 | 19.5 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 6.4 | 32.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 6.1 | 24.4 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 5.5 | 171.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 5.5 | 27.6 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 5.5 | 126.6 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 5.5 | 114.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 5.4 | 37.9 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 5.3 | 42.7 | GO:0015846 | polyamine transport(GO:0015846) |
| 5.3 | 52.8 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 5.2 | 15.5 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 5.1 | 76.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 4.9 | 14.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 4.9 | 63.2 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 4.6 | 27.4 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 4.5 | 54.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 4.4 | 39.9 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 4.2 | 20.9 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 4.1 | 536.3 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 4.1 | 12.3 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 4.1 | 16.2 | GO:0072423 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 4.0 | 12.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 4.0 | 12.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 3.9 | 23.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 3.8 | 138.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 3.8 | 26.6 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 3.8 | 11.4 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 3.6 | 18.1 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 3.6 | 57.6 | GO:0000338 | protein deneddylation(GO:0000338) |
| 3.5 | 41.9 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 3.5 | 38.4 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 3.4 | 17.2 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 3.4 | 60.5 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 3.3 | 9.9 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 3.3 | 9.8 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 3.2 | 12.8 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 3.2 | 19.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 3.2 | 9.5 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 3.1 | 9.3 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 3.0 | 60.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 3.0 | 29.8 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 3.0 | 17.8 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 2.9 | 11.7 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 2.8 | 8.4 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 2.8 | 11.2 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 2.7 | 13.7 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 2.7 | 18.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 2.7 | 13.3 | GO:0072642 | interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 2.6 | 2.6 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 2.5 | 32.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 2.3 | 162.5 | GO:0007077 | mitotic nuclear envelope disassembly(GO:0007077) |
| 2.3 | 7.0 | GO:0048382 | mesendoderm development(GO:0048382) |
| 2.3 | 25.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 2.3 | 13.8 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 2.3 | 18.2 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 2.3 | 9.1 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 2.2 | 15.7 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 2.2 | 37.6 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 2.2 | 28.7 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 2.2 | 15.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 2.1 | 45.0 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 2.1 | 8.4 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 2.1 | 12.4 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 2.1 | 45.5 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 2.1 | 18.5 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 2.0 | 62.3 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 2.0 | 9.8 | GO:0051182 | coenzyme transport(GO:0051182) |
| 2.0 | 5.9 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 1.9 | 29.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 1.9 | 40.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 1.9 | 30.8 | GO:0061684 | chaperone-mediated autophagy(GO:0061684) regulation of chaperone-mediated autophagy(GO:1904714) |
| 1.8 | 7.4 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 1.8 | 36.6 | GO:0002021 | response to dietary excess(GO:0002021) |
| 1.8 | 27.3 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 1.8 | 12.6 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 1.8 | 42.9 | GO:0097286 | iron ion import(GO:0097286) |
| 1.8 | 174.9 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 1.8 | 7.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 1.7 | 26.1 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 1.7 | 17.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 1.7 | 25.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 1.7 | 28.3 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 1.6 | 11.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 1.6 | 9.8 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 1.6 | 6.4 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 1.6 | 6.4 | GO:0019230 | proprioception(GO:0019230) |
| 1.6 | 18.7 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 1.6 | 9.3 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 1.5 | 9.3 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 1.5 | 29.1 | GO:0050872 | positive regulation of chromatin binding(GO:0035563) white fat cell differentiation(GO:0050872) |
| 1.5 | 10.5 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 1.4 | 15.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 1.4 | 39.2 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 1.4 | 5.5 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 1.4 | 43.9 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 1.4 | 12.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 1.4 | 39.6 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 1.3 | 12.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 1.3 | 6.5 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 1.3 | 42.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 1.3 | 35.6 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 1.3 | 12.6 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 1.3 | 8.8 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 1.2 | 6.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 1.2 | 45.6 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 1.2 | 130.4 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 1.2 | 4.8 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 1.2 | 25.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 1.2 | 1.2 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 1.2 | 20.2 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 1.2 | 16.5 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 1.2 | 39.0 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 1.2 | 4.6 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 1.1 | 17.1 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 1.1 | 12.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 1.1 | 3.4 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 1.1 | 82.8 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 1.1 | 5.6 | GO:2000035 | positive regulation of somatic stem cell population maintenance(GO:1904674) regulation of stem cell division(GO:2000035) |
| 1.1 | 45.6 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 1.1 | 13.1 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 1.1 | 4.4 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) positive regulation of Schwann cell differentiation(GO:0014040) response to cobalamin(GO:0033590) |
| 1.1 | 6.5 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 1.1 | 10.9 | GO:0016236 | macroautophagy(GO:0016236) |
| 1.1 | 72.6 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 1.0 | 4.2 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 1.0 | 50.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 1.0 | 29.7 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 1.0 | 32.6 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 1.0 | 8.9 | GO:0006954 | inflammatory response(GO:0006954) |
| 1.0 | 3.8 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 1.0 | 4.8 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.9 | 47.9 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.9 | 11.6 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.9 | 4.4 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.9 | 3.4 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.8 | 3.4 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.8 | 67.7 | GO:0048208 | vesicle coating(GO:0006901) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.8 | 12.5 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.8 | 31.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.8 | 13.9 | GO:0031935 | regulation of chromatin silencing(GO:0031935) |
| 0.8 | 17.6 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.8 | 7.9 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.8 | 10.8 | GO:0048757 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.8 | 8.5 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.8 | 31.4 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.8 | 52.5 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 0.7 | 34.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.7 | 2.9 | GO:1901805 | glucosylceramide catabolic process(GO:0006680) beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.7 | 12.5 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.7 | 9.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.7 | 30.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.7 | 4.2 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.7 | 10.2 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.7 | 53.5 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.7 | 36.1 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.7 | 7.8 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.6 | 1.9 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.6 | 12.6 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.6 | 27.0 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.6 | 1.2 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.6 | 20.8 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.6 | 3.7 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.6 | 4.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.6 | 3.5 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.6 | 26.8 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.5 | 26.6 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.5 | 7.3 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.5 | 35.8 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.5 | 22.9 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.5 | 13.6 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.5 | 15.3 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.5 | 10.2 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.5 | 33.6 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.5 | 145.3 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.5 | 20.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.5 | 6.8 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.5 | 11.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.4 | 9.8 | GO:0007567 | parturition(GO:0007567) |
| 0.4 | 17.2 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.4 | 6.9 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.4 | 36.2 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.4 | 29.6 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.3 | 14.2 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.3 | 6.7 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.3 | 28.1 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.3 | 1.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.3 | 8.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 14.8 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.3 | 0.8 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.3 | 30.8 | GO:0048675 | axon extension(GO:0048675) |
| 0.3 | 2.5 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.3 | 0.8 | GO:0032912 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of transforming growth factor beta2 production(GO:0032912) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.2 | 2.2 | GO:0015791 | polyol transport(GO:0015791) |
| 0.2 | 9.1 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.2 | 15.8 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.2 | 11.4 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.2 | 8.9 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.2 | 4.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.2 | 11.9 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 3.8 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.1 | 2.8 | GO:1901796 | regulation of signal transduction by p53 class mediator(GO:1901796) |
| 0.1 | 10.7 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 0.4 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 9.0 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 1.7 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.1 | 1.1 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 | 1.2 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 5.5 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 3.5 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.7 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.0 | 0.2 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 1.0 | GO:0016310 | phosphorylation(GO:0016310) |
| 0.0 | 2.1 | GO:0017038 | protein import(GO:0017038) |
| 0.0 | 1.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 2.7 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.9 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 2.3 | GO:0002576 | platelet degranulation(GO:0002576) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 35.9 | 143.6 | GO:1990423 | RZZ complex(GO:1990423) |
| 32.2 | 96.6 | GO:0034455 | t-UTP complex(GO:0034455) |
| 31.7 | 126.6 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 26.5 | 265.5 | GO:0000796 | condensin complex(GO:0000796) |
| 25.5 | 101.9 | GO:0031417 | NatC complex(GO:0031417) |
| 21.2 | 381.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 15.7 | 94.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 14.5 | 72.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 14.2 | 255.3 | GO:0034709 | methylosome(GO:0034709) |
| 14.0 | 42.1 | GO:0030689 | Noc complex(GO:0030689) |
| 14.0 | 83.7 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 12.0 | 48.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 9.8 | 127.9 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 8.6 | 128.6 | GO:0044754 | autolysosome(GO:0044754) |
| 8.1 | 154.7 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 8.0 | 39.9 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 7.7 | 781.6 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 7.4 | 51.8 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 7.3 | 58.1 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 7.1 | 213.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 7.0 | 56.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 6.9 | 27.6 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 6.7 | 74.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 6.5 | 71.6 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 6.3 | 234.6 | GO:0016592 | mediator complex(GO:0016592) |
| 6.3 | 31.4 | GO:0032021 | NELF complex(GO:0032021) |
| 6.3 | 37.7 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 6.1 | 42.9 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 6.0 | 42.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 5.9 | 41.4 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 5.5 | 27.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 5.5 | 27.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 5.4 | 37.9 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 5.4 | 59.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 5.3 | 79.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 5.2 | 15.7 | GO:0005826 | actomyosin contractile ring(GO:0005826) GID complex(GO:0034657) |
| 5.2 | 124.7 | GO:0046930 | pore complex(GO:0046930) |
| 5.0 | 35.0 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 4.9 | 14.8 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 4.8 | 86.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 4.7 | 98.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 4.7 | 41.9 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 4.5 | 18.1 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 4.2 | 55.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 4.2 | 12.6 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 4.2 | 112.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 4.1 | 37.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 4.1 | 12.3 | GO:0031213 | RSF complex(GO:0031213) |
| 4.1 | 24.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 4.0 | 59.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 3.9 | 11.7 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 3.7 | 18.5 | GO:0035061 | interchromatin granule(GO:0035061) |
| 3.7 | 14.8 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 3.5 | 60.0 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 3.5 | 379.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 3.5 | 35.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 3.5 | 31.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 3.4 | 85.1 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 3.3 | 26.7 | GO:0000322 | storage vacuole(GO:0000322) |
| 3.3 | 45.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 3.3 | 19.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 3.1 | 24.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 3.0 | 32.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 3.0 | 109.4 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 2.9 | 11.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 2.9 | 17.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 2.8 | 94.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 2.8 | 55.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 2.8 | 239.8 | GO:0005840 | ribosome(GO:0005840) |
| 2.7 | 43.8 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 2.7 | 134.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 2.6 | 186.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 2.6 | 18.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 2.6 | 23.0 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 2.4 | 14.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 2.4 | 9.7 | GO:0071817 | MMXD complex(GO:0071817) |
| 2.4 | 115.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 2.4 | 78.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 2.4 | 26.1 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 2.4 | 42.7 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 2.2 | 29.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 2.2 | 204.5 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 2.2 | 37.6 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 2.2 | 4.4 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 2.1 | 40.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 2.1 | 33.0 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 2.0 | 9.9 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 1.9 | 48.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 1.9 | 101.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 1.8 | 5.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 1.8 | 113.1 | GO:0015030 | Cajal body(GO:0015030) |
| 1.8 | 5.3 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 1.7 | 10.4 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 1.7 | 17.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 1.7 | 20.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 1.7 | 8.5 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 1.6 | 40.9 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 1.6 | 48.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 1.6 | 4.7 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 1.5 | 72.7 | GO:0002102 | podosome(GO:0002102) |
| 1.5 | 75.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 1.5 | 68.5 | GO:0043034 | costamere(GO:0043034) |
| 1.5 | 24.4 | GO:0030426 | growth cone(GO:0030426) |
| 1.5 | 12.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 1.5 | 39.1 | GO:0005844 | polysome(GO:0005844) |
| 1.5 | 7.5 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 1.5 | 28.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 1.5 | 5.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 1.4 | 11.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 1.4 | 109.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 1.4 | 235.8 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 1.4 | 13.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 1.4 | 8.3 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 1.4 | 12.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 1.4 | 8.1 | GO:0090543 | Flemming body(GO:0090543) |
| 1.3 | 31.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 1.2 | 3.7 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 1.2 | 13.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 1.1 | 15.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 1.1 | 2.2 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 1.1 | 43.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 1.1 | 11.6 | GO:0005685 | U1 snRNP(GO:0005685) |
| 1.1 | 35.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 1.0 | 9.4 | GO:0032039 | integrator complex(GO:0032039) |
| 1.0 | 106.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 1.0 | 24.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 1.0 | 12.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 1.0 | 9.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 1.0 | 11.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 1.0 | 11.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.9 | 5.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.9 | 70.8 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.8 | 10.8 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.8 | 78.8 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.8 | 7.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.8 | 104.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.8 | 311.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.8 | 61.7 | GO:0031970 | organelle envelope lumen(GO:0031970) |
| 0.8 | 3.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.8 | 12.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.7 | 5.7 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.7 | 11.3 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.7 | 7.1 | GO:0070187 | telosome(GO:0070187) |
| 0.7 | 266.9 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.6 | 53.6 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.6 | 3.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.5 | 21.1 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.5 | 6.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.5 | 4.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.5 | 9.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.5 | 133.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.4 | 4.4 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.4 | 6.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.4 | 6.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.4 | 7.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.4 | 1.9 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.4 | 31.9 | GO:0005814 | centriole(GO:0005814) |
| 0.4 | 6.1 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.4 | 18.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.3 | 3.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.3 | 7.5 | GO:0045202 | synapse(GO:0045202) |
| 0.3 | 12.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.3 | 12.4 | GO:0032155 | cell division site(GO:0032153) cell division site part(GO:0032155) |
| 0.3 | 93.9 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.2 | 17.2 | GO:0005874 | microtubule(GO:0005874) |
| 0.2 | 30.9 | GO:0005938 | cell cortex(GO:0005938) |
| 0.2 | 138.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.2 | 16.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.2 | 12.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 6.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 36.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 5.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 4.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 23.8 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.1 | 10.9 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 2.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.1 | GO:0098793 | presynapse(GO:0098793) |
| 0.1 | 30.9 | GO:0030425 | dendrite(GO:0030425) |
| 0.1 | 9.5 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
| 0.1 | 5.9 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 1.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 1.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 21.5 | GO:0042175 | nuclear outer membrane-endoplasmic reticulum membrane network(GO:0042175) |
| 0.0 | 28.4 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 35.5 | 213.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 31.7 | 126.6 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 30.0 | 150.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 23.4 | 187.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 21.8 | 109.0 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 21.3 | 42.7 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 20.6 | 144.4 | GO:0050733 | RS domain binding(GO:0050733) |
| 19.6 | 195.6 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 18.5 | 55.5 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 17.9 | 71.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 16.7 | 766.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 16.0 | 175.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 15.4 | 416.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 14.9 | 29.7 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 14.8 | 44.4 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 14.3 | 42.9 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 13.5 | 94.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 13.3 | 106.5 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 12.8 | 38.4 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 12.1 | 109.2 | GO:0015288 | porin activity(GO:0015288) |
| 12.1 | 72.6 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 10.7 | 32.1 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 10.5 | 41.9 | GO:0036033 | mediator complex binding(GO:0036033) |
| 10.2 | 30.7 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 9.9 | 49.7 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 9.5 | 47.6 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 8.9 | 44.3 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 8.3 | 75.0 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 8.2 | 115.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 8.0 | 39.9 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 7.8 | 23.5 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 7.8 | 39.2 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 7.6 | 53.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 7.0 | 20.9 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 7.0 | 41.7 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 6.7 | 40.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 6.6 | 19.9 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 6.6 | 72.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 6.5 | 103.2 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 6.4 | 32.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 6.3 | 56.4 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 6.2 | 37.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 6.1 | 84.9 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 6.0 | 23.8 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 5.7 | 22.9 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 5.2 | 15.5 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 5.1 | 25.4 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 5.0 | 146.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 5.0 | 169.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 4.9 | 14.8 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 4.9 | 14.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 4.8 | 164.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 4.8 | 129.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 4.7 | 18.7 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 4.6 | 27.6 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 4.4 | 35.4 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 4.2 | 12.6 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 4.2 | 29.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 4.1 | 37.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 4.1 | 24.5 | GO:0046979 | TAP2 binding(GO:0046979) |
| 3.8 | 11.4 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 3.4 | 78.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 3.4 | 17.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 3.3 | 666.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 3.3 | 13.3 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 3.3 | 9.9 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 3.3 | 9.8 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 3.2 | 12.9 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 3.2 | 19.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 3.1 | 52.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 3.1 | 9.3 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 3.1 | 46.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 3.0 | 12.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 3.0 | 35.7 | GO:0089720 | caspase binding(GO:0089720) |
| 2.9 | 111.9 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 2.8 | 138.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 2.8 | 11.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 2.8 | 19.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 2.6 | 13.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 2.6 | 91.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 2.6 | 10.2 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 2.5 | 7.5 | GO:0034046 | poly(G) binding(GO:0034046) |
| 2.5 | 62.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 2.5 | 17.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 2.4 | 61.4 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 2.3 | 18.5 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 2.2 | 31.0 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 2.2 | 39.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 2.1 | 10.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 2.0 | 37.6 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 2.0 | 81.2 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 1.9 | 94.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 1.9 | 122.3 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 1.9 | 49.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 1.8 | 31.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 1.8 | 7.3 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 1.8 | 9.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 1.8 | 197.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 1.8 | 116.2 | GO:0031491 | nucleosome binding(GO:0031491) |
| 1.7 | 35.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 1.7 | 6.8 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 1.7 | 8.5 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 1.7 | 8.4 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 1.7 | 6.7 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 1.6 | 9.9 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 1.6 | 4.8 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 1.6 | 12.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 1.6 | 12.4 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 1.5 | 4.6 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 1.5 | 22.9 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 1.5 | 19.5 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 1.4 | 37.5 | GO:0030515 | snoRNA binding(GO:0030515) |
| 1.4 | 4.2 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 1.4 | 21.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 1.4 | 13.9 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 1.4 | 4.2 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 1.4 | 12.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 1.4 | 64.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 1.3 | 95.2 | GO:0009055 | electron carrier activity(GO:0009055) |
| 1.3 | 7.9 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 1.3 | 14.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 1.3 | 68.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 1.3 | 33.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 1.3 | 11.4 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 1.3 | 7.5 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 1.2 | 16.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 1.2 | 13.0 | GO:0039706 | co-receptor binding(GO:0039706) |
| 1.2 | 25.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 1.2 | 26.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 1.1 | 13.6 | GO:0031386 | protein tag(GO:0031386) |
| 1.1 | 4.4 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 1.1 | 33.0 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 1.1 | 17.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 1.0 | 53.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 1.0 | 14.0 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 1.0 | 113.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 1.0 | 40.7 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.9 | 53.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.9 | 23.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.9 | 5.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.8 | 15.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.8 | 6.4 | GO:0016301 | kinase activity(GO:0016301) |
| 0.8 | 2.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.8 | 20.3 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.7 | 2.9 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.7 | 4.4 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.7 | 46.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.7 | 65.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.7 | 9.3 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.7 | 11.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.6 | 18.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.6 | 22.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.6 | 14.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.6 | 76.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.6 | 13.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.6 | 6.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.5 | 44.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.5 | 45.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.5 | 15.4 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.5 | 14.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.5 | 12.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.5 | 9.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.5 | 37.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.5 | 72.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.5 | 3.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.5 | 5.3 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.5 | 651.3 | GO:0003723 | RNA binding(GO:0003723) |
| 0.4 | 115.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.4 | 1.3 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.4 | 8.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.4 | 34.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.4 | 38.2 | GO:0005525 | GTP binding(GO:0005525) |
| 0.4 | 2.2 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.3 | 8.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 2.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.3 | 93.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.3 | 6.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.3 | 13.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.3 | 1.1 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.2 | 4.2 | GO:0097200 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 17.5 | GO:0042393 | histone binding(GO:0042393) |
| 0.2 | 6.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 2.4 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.1 | 3.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 3.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 86.2 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.1 | 10.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.8 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.1 | 0.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 3.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 3.5 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 0.6 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 1.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 32.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 6.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 2.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.2 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.0 | 671.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 5.1 | 167.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 4.9 | 317.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 4.5 | 18.1 | PID BARD1 PATHWAY | BARD1 signaling events |
| 4.0 | 239.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 3.1 | 91.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 2.9 | 145.5 | PID ATR PATHWAY | ATR signaling pathway |
| 2.4 | 82.5 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 2.4 | 78.7 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 2.1 | 84.9 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 2.1 | 220.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 2.0 | 108.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 1.9 | 95.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 1.5 | 35.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 1.3 | 53.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 1.2 | 81.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 1.1 | 89.0 | PID E2F PATHWAY | E2F transcription factor network |
| 1.1 | 23.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 1.0 | 38.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 1.0 | 15.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 1.0 | 12.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.9 | 86.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.9 | 32.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.9 | 3.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.9 | 6.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.8 | 50.2 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.8 | 22.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.8 | 25.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.8 | 25.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.7 | 42.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.5 | 29.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.4 | 28.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.4 | 30.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.4 | 15.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.3 | 17.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.3 | 11.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.2 | 10.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.2 | 14.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 2.6 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.2 | 8.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.2 | 7.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 44.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 2.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 9.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.2 | 6.7 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.2 | 8.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 8.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 6.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 6.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 18.7 | 692.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 18.4 | 424.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 10.8 | 303.1 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 10.2 | 183.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 9.1 | 145.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 7.1 | 537.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 6.3 | 94.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 6.0 | 72.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 4.8 | 126.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 4.6 | 480.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 4.5 | 245.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 4.4 | 35.0 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 4.3 | 12.9 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 4.1 | 86.9 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 4.1 | 127.9 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 4.1 | 190.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 4.0 | 145.1 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 3.8 | 37.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 3.6 | 32.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 3.5 | 51.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 3.4 | 64.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 3.3 | 274.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 3.3 | 253.9 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 3.2 | 44.8 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 3.2 | 12.8 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 3.0 | 108.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 3.0 | 29.8 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 2.9 | 72.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 2.8 | 5.5 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 2.8 | 146.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 2.6 | 82.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 2.4 | 23.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 2.2 | 60.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 2.2 | 17.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 2.1 | 59.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 2.1 | 38.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 1.9 | 23.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 1.9 | 123.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 1.9 | 161.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 1.9 | 44.9 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 1.8 | 56.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 1.7 | 69.9 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 1.6 | 9.8 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 1.5 | 38.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 1.5 | 27.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 1.3 | 42.9 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 1.2 | 28.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 1.1 | 11.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 1.1 | 13.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 1.1 | 74.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 1.0 | 28.3 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 1.0 | 9.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 1.0 | 18.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 1.0 | 23.4 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 1.0 | 14.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 1.0 | 18.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.9 | 15.0 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.9 | 23.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.9 | 99.8 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.8 | 13.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.7 | 26.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.7 | 20.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.7 | 22.9 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.7 | 86.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.7 | 40.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.6 | 7.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.6 | 15.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.6 | 15.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.5 | 14.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.5 | 34.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.4 | 29.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.4 | 8.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.4 | 20.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.4 | 10.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.4 | 3.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.4 | 7.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.4 | 9.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.3 | 6.9 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.3 | 58.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.3 | 11.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.3 | 40.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.3 | 2.7 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.3 | 4.4 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.3 | 5.9 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.3 | 5.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.3 | 4.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 5.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 4.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.2 | 4.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 6.8 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 8.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 2.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 3.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 4.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 2.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 5.4 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |