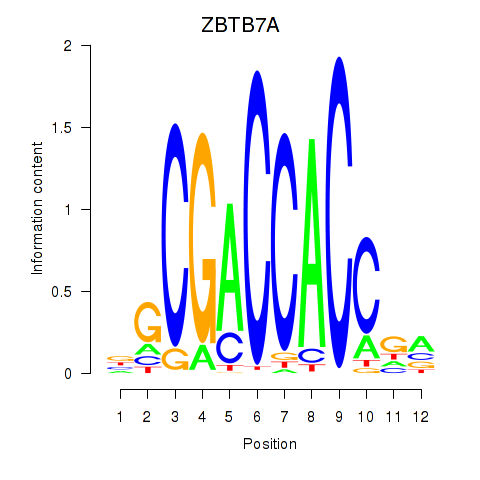
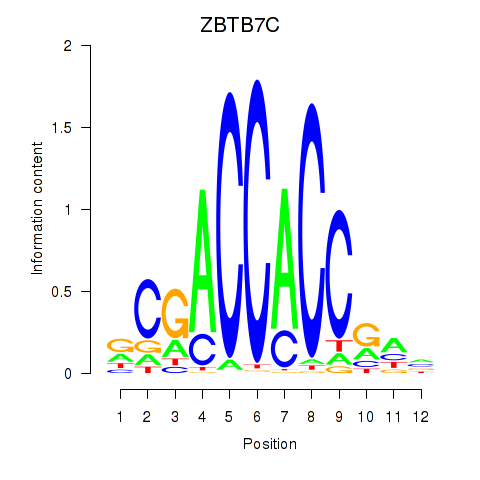
Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for ZBTB7A_ZBTB7C
Z-value: 1.09
Transcription factors associated with ZBTB7A_ZBTB7C
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB7A
|
ENSG00000178951.4 | zinc finger and BTB domain containing 7A |
|
ZBTB7C
|
ENSG00000184828.5 | zinc finger and BTB domain containing 7C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB7A | hg19_v2_chr19_-_4066890_4066943 | 0.38 | 4.1e-09 | Click! |
| ZBTB7C | hg19_v2_chr18_-_45663666_45663732, hg19_v2_chr18_-_45935663_45935793 | 0.03 | 6.8e-01 | Click! |
Activity profile of ZBTB7A_ZBTB7C motif
Sorted Z-values of ZBTB7A_ZBTB7C motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_132112907 | 38.52 |
ENST00000458488.2
|
SEPT8
|
septin 8 |
| chr5_-_132112921 | 38.06 |
ENST00000378721.4
ENST00000378701.1 |
SEPT8
|
septin 8 |
| chr1_+_169075554 | 21.05 |
ENST00000367815.4
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr19_+_35634146 | 19.57 |
ENST00000586063.1
ENST00000270310.2 ENST00000588265.1 |
FXYD7
|
FXYD domain containing ion transport regulator 7 |
| chr14_+_29236269 | 18.22 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr15_-_64126084 | 17.96 |
ENST00000560316.1
ENST00000443617.2 ENST00000560462.1 ENST00000558532.1 ENST00000561400.1 |
HERC1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr7_-_94285402 | 17.86 |
ENST00000428696.2
ENST00000445866.2 |
SGCE
|
sarcoglycan, epsilon |
| chr4_+_41258786 | 17.86 |
ENST00000503431.1
ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr11_-_66115032 | 17.46 |
ENST00000311181.4
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 |
| chr17_+_40834580 | 16.77 |
ENST00000264638.4
|
CNTNAP1
|
contactin associated protein 1 |
| chr20_+_36149602 | 16.56 |
ENST00000062104.2
ENST00000346199.2 |
NNAT
|
neuronatin |
| chr5_+_139493665 | 16.24 |
ENST00000331327.3
|
PURA
|
purine-rich element binding protein A |
| chr5_-_132113036 | 15.75 |
ENST00000378706.1
|
SEPT8
|
septin 8 |
| chr20_+_44034804 | 15.51 |
ENST00000357275.2
ENST00000372720.3 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr7_-_94285472 | 14.71 |
ENST00000437425.2
ENST00000447873.1 ENST00000415788.2 |
SGCE
|
sarcoglycan, epsilon |
| chr14_+_100150622 | 14.58 |
ENST00000261835.3
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1 |
| chr16_+_15528332 | 14.29 |
ENST00000566490.1
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr20_+_44034676 | 14.02 |
ENST00000372723.3
ENST00000372722.3 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr11_-_117747607 | 13.77 |
ENST00000540359.1
ENST00000539526.1 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr12_+_13197218 | 13.66 |
ENST00000197268.8
|
KIAA1467
|
KIAA1467 |
| chr7_-_94285511 | 13.62 |
ENST00000265735.7
|
SGCE
|
sarcoglycan, epsilon |
| chr11_-_117747434 | 13.34 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr16_-_19896220 | 13.02 |
ENST00000562469.1
ENST00000300571.2 |
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B |
| chr6_-_46459675 | 12.98 |
ENST00000306764.7
|
RCAN2
|
regulator of calcineurin 2 |
| chr5_+_76506706 | 11.62 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr6_-_122792919 | 11.18 |
ENST00000339697.4
|
SERINC1
|
serine incorporator 1 |
| chr17_+_43972010 | 9.53 |
ENST00000334239.8
ENST00000446361.3 |
MAPT
|
microtubule-associated protein tau |
| chr3_-_33759541 | 9.40 |
ENST00000468888.2
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr5_-_132113083 | 9.26 |
ENST00000296873.7
|
SEPT8
|
septin 8 |
| chrX_-_51812268 | 9.23 |
ENST00000486010.1
ENST00000497164.1 ENST00000360134.6 ENST00000485287.1 ENST00000335504.5 ENST00000431659.1 |
MAGED4B
|
melanoma antigen family D, 4B |
| chr9_-_123476612 | 8.83 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr11_-_1330834 | 8.80 |
ENST00000525159.1
ENST00000317204.6 ENST00000542915.1 ENST00000527938.1 ENST00000530541.1 ENST00000263646.7 |
TOLLIP
|
toll interacting protein |
| chr3_-_33759699 | 8.43 |
ENST00000399362.4
ENST00000359576.5 ENST00000307312.7 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr5_-_132113559 | 8.39 |
ENST00000448933.1
|
SEPT8
|
septin 8 |
| chr14_-_22005062 | 8.37 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr17_-_1532106 | 8.01 |
ENST00000301335.5
ENST00000382147.4 |
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr7_-_51384451 | 7.76 |
ENST00000441453.1
ENST00000265136.7 ENST00000395542.2 ENST00000395540.2 |
COBL
|
cordon-bleu WH2 repeat protein |
| chr1_-_95007193 | 6.77 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr9_+_74526384 | 6.74 |
ENST00000334731.2
ENST00000377031.3 |
C9orf85
|
chromosome 9 open reading frame 85 |
| chr22_+_31031639 | 6.62 |
ENST00000343605.4
ENST00000300385.8 |
SLC35E4
|
solute carrier family 35, member E4 |
| chr11_+_133938820 | 6.38 |
ENST00000299106.4
ENST00000529443.2 |
JAM3
|
junctional adhesion molecule 3 |
| chr5_-_443239 | 6.37 |
ENST00000408966.2
|
C5orf55
|
chromosome 5 open reading frame 55 |
| chrX_+_133507327 | 6.32 |
ENST00000332070.3
ENST00000394292.1 ENST00000370799.1 ENST00000416404.2 |
PHF6
|
PHD finger protein 6 |
| chr5_+_178286925 | 6.04 |
ENST00000322434.3
|
ZNF354B
|
zinc finger protein 354B |
| chr11_+_133938955 | 6.02 |
ENST00000534549.1
ENST00000441717.3 |
JAM3
|
junctional adhesion molecule 3 |
| chr5_-_138210977 | 5.85 |
ENST00000274711.6
ENST00000521094.2 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr8_-_144242020 | 5.47 |
ENST00000414417.2
|
LY6H
|
lymphocyte antigen 6 complex, locus H |
| chr6_-_36953833 | 5.46 |
ENST00000538808.1
ENST00000460219.1 ENST00000373616.5 ENST00000373627.5 |
MTCH1
|
mitochondrial carrier 1 |
| chr13_+_96204961 | 5.41 |
ENST00000299339.2
|
CLDN10
|
claudin 10 |
| chr15_-_83876758 | 5.34 |
ENST00000299633.4
|
HDGFRP3
|
Hepatoma-derived growth factor-related protein 3 |
| chr14_+_29234870 | 5.33 |
ENST00000382535.3
|
FOXG1
|
forkhead box G1 |
| chr15_-_23932437 | 5.08 |
ENST00000331837.4
|
NDN
|
necdin, melanoma antigen (MAGE) family member |
| chr8_-_144241664 | 4.91 |
ENST00000342752.4
|
LY6H
|
lymphocyte antigen 6 complex, locus H |
| chr8_-_144241432 | 4.90 |
ENST00000430474.2
|
LY6H
|
lymphocyte antigen 6 complex, locus H |
| chr1_-_151431909 | 4.73 |
ENST00000361398.3
ENST00000271715.2 |
POGZ
|
pogo transposable element with ZNF domain |
| chr17_-_8534031 | 4.72 |
ENST00000411957.1
ENST00000396239.1 ENST00000379980.4 |
MYH10
|
myosin, heavy chain 10, non-muscle |
| chr17_-_5372271 | 4.72 |
ENST00000225296.3
|
DHX33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr17_-_8534067 | 4.59 |
ENST00000360416.3
ENST00000269243.4 |
MYH10
|
myosin, heavy chain 10, non-muscle |
| chr20_-_36156293 | 4.45 |
ENST00000373537.2
ENST00000414542.2 |
BLCAP
|
bladder cancer associated protein |
| chr4_+_156588350 | 4.42 |
ENST00000296518.7
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr8_+_9413410 | 4.36 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr12_-_42631529 | 4.36 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr20_-_36156125 | 4.33 |
ENST00000397135.1
ENST00000397137.1 |
BLCAP
|
bladder cancer associated protein |
| chr2_+_136289030 | 4.27 |
ENST00000409478.1
ENST00000264160.4 ENST00000329971.3 ENST00000438014.1 |
R3HDM1
|
R3H domain containing 1 |
| chr14_-_22005018 | 4.27 |
ENST00000546363.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr2_-_152118352 | 4.26 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr3_+_15468862 | 4.19 |
ENST00000396842.2
|
EAF1
|
ELL associated factor 1 |
| chr2_+_25016282 | 4.13 |
ENST00000260662.1
|
CENPO
|
centromere protein O |
| chr2_+_198669365 | 4.11 |
ENST00000428675.1
|
PLCL1
|
phospholipase C-like 1 |
| chr3_+_108308513 | 4.10 |
ENST00000361582.3
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr5_+_149109825 | 4.01 |
ENST00000360453.4
ENST00000394320.3 ENST00000309241.5 |
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr2_+_25015968 | 3.79 |
ENST00000380834.2
ENST00000473706.1 |
CENPO
|
centromere protein O |
| chr5_-_114515734 | 3.77 |
ENST00000514154.1
ENST00000282369.3 |
TRIM36
|
tripartite motif containing 36 |
| chr17_-_4890919 | 3.73 |
ENST00000572543.1
ENST00000381311.5 ENST00000348066.3 ENST00000358183.4 |
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr3_-_149470229 | 3.56 |
ENST00000473414.1
|
COMMD2
|
COMM domain containing 2 |
| chr3_-_15469006 | 3.55 |
ENST00000443029.1
ENST00000383790.3 ENST00000383789.5 |
METTL6
|
methyltransferase like 6 |
| chr8_-_38853990 | 3.45 |
ENST00000456845.2
ENST00000397070.2 ENST00000517872.1 ENST00000412303.1 ENST00000456397.2 |
TM2D2
|
TM2 domain containing 2 |
| chr8_-_101964231 | 3.40 |
ENST00000521309.1
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr8_-_101964265 | 3.23 |
ENST00000395958.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr12_+_57943781 | 3.13 |
ENST00000455537.2
ENST00000286452.5 |
KIF5A
|
kinesin family member 5A |
| chr8_+_26240414 | 3.01 |
ENST00000380629.2
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr1_-_43205811 | 2.99 |
ENST00000372539.3
ENST00000296387.1 ENST00000539749.1 |
CLDN19
|
claudin 19 |
| chr17_-_7137582 | 2.89 |
ENST00000575756.1
ENST00000575458.1 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr6_+_87865262 | 2.87 |
ENST00000369577.3
ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chrX_-_119603138 | 2.86 |
ENST00000200639.4
ENST00000371335.4 ENST00000538785.1 ENST00000434600.2 |
LAMP2
|
lysosomal-associated membrane protein 2 |
| chr19_+_5904866 | 2.80 |
ENST00000339485.3
|
VMAC
|
vimentin-type intermediate filament associated coiled-coil protein |
| chr14_-_102771462 | 2.70 |
ENST00000522874.1
|
MOK
|
MOK protein kinase |
| chr9_+_132934835 | 2.68 |
ENST00000372398.3
|
NCS1
|
neuronal calcium sensor 1 |
| chrX_-_153141302 | 2.68 |
ENST00000361699.4
ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM
|
L1 cell adhesion molecule |
| chr4_+_39184024 | 2.67 |
ENST00000399820.3
ENST00000509560.1 ENST00000512112.1 ENST00000288634.7 ENST00000506503.1 |
WDR19
|
WD repeat domain 19 |
| chr9_-_139094988 | 2.65 |
ENST00000371746.3
|
LHX3
|
LIM homeobox 3 |
| chr5_-_140070897 | 2.63 |
ENST00000448240.1
ENST00000438307.2 ENST00000415192.2 ENST00000457527.2 ENST00000307633.3 ENST00000507746.1 ENST00000431330.2 |
HARS
|
histidyl-tRNA synthetase |
| chr7_+_66093851 | 2.56 |
ENST00000275532.3
|
KCTD7
|
potassium channel tetramerization domain containing 7 |
| chr5_+_175792459 | 2.50 |
ENST00000310389.5
|
ARL10
|
ADP-ribosylation factor-like 10 |
| chr14_-_54955721 | 2.45 |
ENST00000554908.1
|
GMFB
|
glia maturation factor, beta |
| chr19_+_49622646 | 2.35 |
ENST00000334186.4
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr9_-_131790464 | 2.21 |
ENST00000417224.1
ENST00000416629.1 ENST00000372559.1 |
SH3GLB2
|
SH3-domain GRB2-like endophilin B2 |
| chr19_-_5567996 | 2.20 |
ENST00000448587.1
|
TINCR
|
tissue differentiation-inducing non-protein coding RNA |
| chr1_+_167905894 | 2.19 |
ENST00000367843.3
ENST00000432587.2 ENST00000312263.6 |
DCAF6
|
DDB1 and CUL4 associated factor 6 |
| chr17_-_37844267 | 2.18 |
ENST00000579146.1
ENST00000378011.4 ENST00000429199.2 ENST00000300658.4 |
PGAP3
|
post-GPI attachment to proteins 3 |
| chr3_-_100120223 | 2.17 |
ENST00000284320.5
|
TOMM70A
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) |
| chr14_+_70233810 | 2.14 |
ENST00000394366.2
ENST00000553548.1 ENST00000553369.1 ENST00000557154.1 ENST00000451983.2 ENST00000553635.1 |
SRSF5
|
serine/arginine-rich splicing factor 5 |
| chr3_+_23986748 | 2.14 |
ENST00000312521.4
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr9_+_131873227 | 2.13 |
ENST00000358994.4
ENST00000455292.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr3_-_164914640 | 2.08 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK-like family, member 3 |
| chr4_-_76598326 | 2.02 |
ENST00000503660.1
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr4_-_16085314 | 2.01 |
ENST00000510224.1
|
PROM1
|
prominin 1 |
| chr14_+_24583836 | 1.99 |
ENST00000559115.1
ENST00000558215.1 ENST00000557810.1 ENST00000561375.1 ENST00000446197.3 ENST00000559796.1 ENST00000560713.1 ENST00000560901.1 ENST00000559382.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr4_-_16085340 | 1.94 |
ENST00000508167.1
|
PROM1
|
prominin 1 |
| chr17_-_4890649 | 1.93 |
ENST00000361571.5
|
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr10_+_70091812 | 1.92 |
ENST00000265866.7
|
HNRNPH3
|
heterogeneous nuclear ribonucleoprotein H3 (2H9) |
| chr14_-_102771516 | 1.78 |
ENST00000524214.1
ENST00000193029.6 ENST00000361847.2 |
MOK
|
MOK protein kinase |
| chr4_-_146101304 | 1.78 |
ENST00000447906.2
|
OTUD4
|
OTU domain containing 4 |
| chr19_+_40697514 | 1.77 |
ENST00000253055.3
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr11_-_63536113 | 1.76 |
ENST00000433688.1
ENST00000546282.2 |
C11orf95
RP11-466C23.4
|
chromosome 11 open reading frame 95 RP11-466C23.4 |
| chr1_-_21616901 | 1.76 |
ENST00000436918.2
ENST00000374893.6 |
ECE1
|
endothelin converting enzyme 1 |
| chr5_-_72861484 | 1.74 |
ENST00000296785.3
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr14_+_59655369 | 1.71 |
ENST00000360909.3
ENST00000351081.1 ENST00000556135.1 |
DAAM1
|
dishevelled associated activator of morphogenesis 1 |
| chr14_-_24584138 | 1.69 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chr19_-_14228541 | 1.65 |
ENST00000590853.1
ENST00000308677.4 |
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha |
| chr22_+_42229100 | 1.64 |
ENST00000361204.4
|
SREBF2
|
sterol regulatory element binding transcription factor 2 |
| chr16_+_84002234 | 1.55 |
ENST00000305202.4
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2 |
| chr17_+_29158962 | 1.54 |
ENST00000321990.4
|
ATAD5
|
ATPase family, AAA domain containing 5 |
| chr11_-_129872712 | 1.51 |
ENST00000358825.5
ENST00000360871.3 ENST00000528746.1 |
PRDM10
|
PR domain containing 10 |
| chrX_+_67913471 | 1.49 |
ENST00000374597.3
|
STARD8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr4_+_72052964 | 1.47 |
ENST00000264485.5
ENST00000425175.1 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr16_-_49315731 | 1.46 |
ENST00000219197.6
|
CBLN1
|
cerebellin 1 precursor |
| chr1_+_25664408 | 1.45 |
ENST00000374358.4
|
TMEM50A
|
transmembrane protein 50A |
| chr12_-_49351148 | 1.43 |
ENST00000398092.4
ENST00000539611.1 |
RP11-302B13.5
ARF3
|
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chrX_-_17878827 | 1.39 |
ENST00000360011.1
|
RAI2
|
retinoic acid induced 2 |
| chr19_-_46296011 | 1.37 |
ENST00000377735.3
ENST00000270223.6 |
DMWD
|
dystrophia myotonica, WD repeat containing |
| chr5_+_443268 | 1.36 |
ENST00000512944.1
|
EXOC3
|
exocyst complex component 3 |
| chr21_-_45079341 | 1.32 |
ENST00000443485.1
ENST00000291560.2 |
HSF2BP
|
heat shock transcription factor 2 binding protein |
| chr8_-_74884511 | 1.29 |
ENST00000518127.1
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr10_+_49514698 | 1.28 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr8_-_101963677 | 1.24 |
ENST00000395956.3
ENST00000395953.2 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr12_-_107487604 | 1.23 |
ENST00000008527.5
|
CRY1
|
cryptochrome 1 (photolyase-like) |
| chr20_-_58508702 | 1.18 |
ENST00000357552.3
ENST00000425931.1 |
SYCP2
|
synaptonemal complex protein 2 |
| chr7_+_43152191 | 1.17 |
ENST00000395891.2
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr5_+_78407602 | 1.14 |
ENST00000274353.5
ENST00000524080.1 |
BHMT
|
betaine--homocysteine S-methyltransferase |
| chr17_+_37844331 | 1.11 |
ENST00000578199.1
ENST00000406381.2 |
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr19_-_56988677 | 1.11 |
ENST00000504904.3
ENST00000292069.6 |
ZNF667
|
zinc finger protein 667 |
| chr17_+_79990058 | 1.08 |
ENST00000584341.1
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chrX_+_135579238 | 1.07 |
ENST00000535601.1
ENST00000448450.1 ENST00000425695.1 |
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr8_-_74884399 | 1.07 |
ENST00000520210.1
ENST00000602840.1 |
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr19_-_59070239 | 1.04 |
ENST00000595957.1
ENST00000253023.3 |
UBE2M
|
ubiquitin-conjugating enzyme E2M |
| chr2_+_242255297 | 1.02 |
ENST00000401990.1
ENST00000407971.1 ENST00000436795.1 ENST00000411484.1 ENST00000434955.1 ENST00000402092.2 ENST00000441533.1 ENST00000443492.1 ENST00000437066.1 ENST00000429791.1 |
SEPT2
|
septin 2 |
| chr20_-_35492048 | 1.02 |
ENST00000237536.4
|
SOGA1
|
suppressor of glucose, autophagy associated 1 |
| chr6_-_16761678 | 1.02 |
ENST00000244769.4
ENST00000436367.1 |
ATXN1
|
ataxin 1 |
| chr1_-_151431647 | 1.02 |
ENST00000368863.2
ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ
|
pogo transposable element with ZNF domain |
| chr6_+_125474939 | 0.98 |
ENST00000527711.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr6_+_125475335 | 0.94 |
ENST00000532429.1
ENST00000534199.1 |
TPD52L1
|
tumor protein D52-like 1 |
| chr19_-_49622348 | 0.91 |
ENST00000408991.2
|
C19orf73
|
chromosome 19 open reading frame 73 |
| chr15_+_27112948 | 0.91 |
ENST00000555060.1
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr1_+_110754094 | 0.87 |
ENST00000369787.3
ENST00000413138.3 ENST00000438661.2 |
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
| chr9_+_35161998 | 0.86 |
ENST00000396787.1
ENST00000378495.3 ENST00000378496.4 |
UNC13B
|
unc-13 homolog B (C. elegans) |
| chr12_+_50355647 | 0.85 |
ENST00000293599.6
|
AQP5
|
aquaporin 5 |
| chr21_-_47648665 | 0.82 |
ENST00000450351.1
ENST00000522411.1 ENST00000356396.4 ENST00000397728.3 ENST00000457828.2 |
LSS
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr22_+_21369316 | 0.68 |
ENST00000413302.2
ENST00000402329.3 ENST00000336296.2 ENST00000401443.1 ENST00000443995.3 |
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr6_-_34113856 | 0.67 |
ENST00000538487.2
|
GRM4
|
glutamate receptor, metabotropic 4 |
| chr15_+_92397051 | 0.67 |
ENST00000424469.2
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1 |
| chrX_+_44732757 | 0.63 |
ENST00000377967.4
ENST00000536777.1 ENST00000382899.4 ENST00000543216.1 |
KDM6A
|
lysine (K)-specific demethylase 6A |
| chr3_+_50126341 | 0.62 |
ENST00000347869.3
ENST00000469838.1 ENST00000404526.2 ENST00000441305.1 |
RBM5
|
RNA binding motif protein 5 |
| chr6_+_126070726 | 0.61 |
ENST00000368364.3
|
HEY2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr19_+_38755042 | 0.61 |
ENST00000301244.7
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr6_+_125474992 | 0.59 |
ENST00000528193.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr4_-_119274121 | 0.58 |
ENST00000296498.3
|
PRSS12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr19_+_39833036 | 0.49 |
ENST00000602243.1
ENST00000598913.1 ENST00000314471.6 |
SAMD4B
|
sterile alpha motif domain containing 4B |
| chr4_-_83351005 | 0.49 |
ENST00000295470.5
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr2_-_97405775 | 0.48 |
ENST00000264963.4
ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L
|
lectin, mannose-binding 2-like |
| chr1_+_167906056 | 0.46 |
ENST00000367840.3
|
DCAF6
|
DDB1 and CUL4 associated factor 6 |
| chrX_-_17879356 | 0.42 |
ENST00000331511.1
ENST00000415486.3 ENST00000545871.1 ENST00000451717.1 |
RAI2
|
retinoic acid induced 2 |
| chr6_-_30043539 | 0.42 |
ENST00000376751.3
ENST00000244360.6 |
RNF39
|
ring finger protein 39 |
| chr20_-_61493115 | 0.41 |
ENST00000335351.3
ENST00000217162.5 |
TCFL5
|
transcription factor-like 5 (basic helix-loop-helix) |
| chr9_-_96717654 | 0.40 |
ENST00000253968.6
|
BARX1
|
BARX homeobox 1 |
| chr4_-_5021164 | 0.39 |
ENST00000506508.1
ENST00000509419.1 ENST00000307746.4 |
CYTL1
|
cytokine-like 1 |
| chr5_-_139283982 | 0.30 |
ENST00000340391.3
|
NRG2
|
neuregulin 2 |
| chr8_-_74884482 | 0.28 |
ENST00000520242.1
ENST00000519082.1 |
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr6_+_157802165 | 0.27 |
ENST00000414563.2
ENST00000359775.5 |
ZDHHC14
|
zinc finger, DHHC-type containing 14 |
| chr20_-_17662705 | 0.25 |
ENST00000455029.2
|
RRBP1
|
ribosome binding protein 1 |
| chr22_+_38035459 | 0.23 |
ENST00000357436.4
|
SH3BP1
|
SH3-domain binding protein 1 |
| chr17_+_79989937 | 0.21 |
ENST00000580965.1
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chr4_+_72053017 | 0.21 |
ENST00000351898.6
|
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr8_-_74884341 | 0.18 |
ENST00000284811.8
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr20_+_37590942 | 0.13 |
ENST00000373325.2
ENST00000252011.3 ENST00000373323.4 |
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr19_+_36203830 | 0.10 |
ENST00000262630.3
|
ZBTB32
|
zinc finger and BTB domain containing 32 |
| chr19_+_38755203 | 0.08 |
ENST00000587090.1
ENST00000454580.3 |
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr5_-_33984786 | 0.05 |
ENST00000296589.4
|
SLC45A2
|
solute carrier family 45, member 2 |
| chr6_-_33290580 | 0.05 |
ENST00000446511.1
ENST00000446403.1 ENST00000414083.2 ENST00000266000.6 ENST00000374542.5 |
DAXX
|
death-domain associated protein |
| chr5_-_33984741 | 0.04 |
ENST00000382102.3
ENST00000509381.1 ENST00000342059.3 ENST00000345083.5 |
SLC45A2
|
solute carrier family 45, member 2 |
| chr16_+_66400533 | 0.04 |
ENST00000341529.3
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB7A_ZBTB7C
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 17.9 | GO:0007412 | axon target recognition(GO:0007412) |
| 5.3 | 21.1 | GO:1903288 | protein transport into plasma membrane raft(GO:0044861) positive regulation of potassium ion import(GO:1903288) |
| 3.1 | 9.3 | GO:0021592 | fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 2.9 | 14.3 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 2.4 | 16.8 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 2.3 | 6.8 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 2.2 | 17.8 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 1.9 | 7.8 | GO:0001757 | somite specification(GO:0001757) |
| 1.9 | 9.5 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 1.7 | 16.6 | GO:0009249 | protein lipoylation(GO:0009249) |
| 1.6 | 16.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 1.5 | 14.6 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 1.5 | 4.4 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 1.4 | 12.4 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 1.3 | 4.0 | GO:2000768 | positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 1.2 | 18.0 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 1.0 | 3.1 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.9 | 11.2 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.8 | 13.0 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.7 | 4.4 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.7 | 2.2 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.7 | 2.0 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.7 | 2.7 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.6 | 7.4 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.6 | 45.1 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.6 | 8.8 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.5 | 7.5 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.5 | 3.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.5 | 2.5 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.5 | 5.5 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.4 | 5.7 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.4 | 2.6 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.4 | 1.8 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.4 | 1.3 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.4 | 1.7 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.4 | 1.6 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.4 | 3.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.3 | 4.0 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.3 | 13.0 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.3 | 4.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.3 | 11.6 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.3 | 1.5 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) positive regulation of long term synaptic depression(GO:1900454) |
| 0.3 | 0.9 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.3 | 8.0 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.3 | 7.9 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 2.9 | GO:1905146 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) lysosomal protein catabolic process(GO:1905146) |
| 0.2 | 2.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 15.3 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.2 | 5.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 0.6 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.2 | 0.6 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.2 | 1.2 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.2 | 5.1 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.2 | 8.8 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.2 | 5.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 1.1 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.2 | 1.3 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.2 | 1.6 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.2 | 0.7 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.2 | 0.8 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 1.1 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 1.9 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.5 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 33.3 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 0.7 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 3.8 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 2.1 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.1 | 1.0 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 2.1 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.1 | 1.2 | GO:0035112 | genitalia morphogenesis(GO:0035112) |
| 0.1 | 12.2 | GO:0021915 | neural tube development(GO:0021915) |
| 0.1 | 22.5 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.1 | 1.8 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 1.0 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 1.7 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 2.2 | GO:0006505 | GPI anchor metabolic process(GO:0006505) GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 21.2 | GO:0007568 | aging(GO:0007568) |
| 0.1 | 4.1 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.9 | GO:0060384 | innervation(GO:0060384) |
| 0.0 | 2.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 1.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 2.7 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.7 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 1.1 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 1.6 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.7 | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway(GO:0060071) regulation of establishment of planar polarity(GO:0090175) |
| 0.0 | 0.9 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 1.2 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.4 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 1.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.8 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.4 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 1.0 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 46.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 2.4 | 29.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 1.9 | 9.5 | GO:0045298 | tubulin complex(GO:0045298) |
| 1.9 | 9.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 1.8 | 17.8 | GO:0045180 | basal cortex(GO:0045180) |
| 1.8 | 8.8 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 1.1 | 16.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 1.1 | 21.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.0 | 7.8 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.8 | 4.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.7 | 2.9 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.4 | 2.7 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.4 | 1.8 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.4 | 12.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.4 | 1.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.4 | 17.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.3 | 1.7 | GO:1990393 | 3M complex(GO:1990393) |
| 0.3 | 29.5 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 6.8 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.3 | 0.9 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.3 | 4.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.3 | 2.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 4.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 3.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 1.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.2 | 2.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 4.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 6.1 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 2.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 4.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 3.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 1.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 12.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 4.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 6.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 8.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 0.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 1.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.4 | GO:0043073 | male germ cell nucleus(GO:0001673) germ cell nucleus(GO:0043073) |
| 0.0 | 0.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 4.0 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 2.0 | GO:0016235 | aggresome(GO:0016235) lateral plasma membrane(GO:0016328) |
| 0.0 | 7.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.7 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 75.7 | GO:0005856 | cytoskeleton(GO:0005856) |
| 0.0 | 0.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 2.4 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 66.9 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.8 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 4.5 | 17.9 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 2.9 | 8.8 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 2.4 | 9.5 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 2.2 | 13.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.3 | 21.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.9 | 11.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.9 | 45.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.7 | 17.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.7 | 2.2 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.7 | 18.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.6 | 4.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.5 | 4.7 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.4 | 1.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.4 | 5.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 13.7 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.4 | 1.5 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.4 | 1.1 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.3 | 4.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.3 | 8.0 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.3 | 4.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.3 | 2.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 11.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.3 | 0.8 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.2 | 105.9 | GO:0005525 | GTP binding(GO:0005525) |
| 0.2 | 0.6 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 1.0 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 2.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 5.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 0.7 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 1.0 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.2 | 7.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 0.6 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 7.5 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 15.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 13.1 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 1.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 1.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 2.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 3.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.6 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 14.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 3.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 4.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 2.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.7 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 1.9 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 2.9 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.8 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 3.7 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 2.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 6.8 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 1.8 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 5.1 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 5.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 4.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 7.5 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.5 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.7 | GO:0008066 | glutamate receptor activity(GO:0008066) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 9.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 14.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.3 | 17.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 12.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 10.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.2 | 7.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 3.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 15.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 1.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 2.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 5.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 2.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.4 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 3.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 14.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.6 | 23.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.5 | 16.1 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.3 | 7.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.3 | 7.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 9.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 10.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 8.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 6.8 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 7.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 3.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 8.4 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 4.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 4.4 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 10.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 2.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 2.1 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 8.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 2.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 4.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 2.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 4.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 4.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 3.3 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |