|
chr11_-_111783919
Show fit
|
88.55 |
ENST00000531198.1
ENST00000533879.1
|
CRYAB
|
crystallin, alpha B
|
|
chr11_-_111783595
Show fit
|
76.97 |
ENST00000528628.1
|
CRYAB
|
crystallin, alpha B
|
|
chr3_+_153839149
Show fit
|
44.41 |
ENST00000465093.1
ENST00000465817.1
|
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26
|
|
chr12_-_6798616
Show fit
|
40.01 |
ENST00000355772.4
ENST00000417772.3
ENST00000396801.3
ENST00000396799.2
|
ZNF384
|
zinc finger protein 384
|
|
chr20_+_36149602
Show fit
|
37.54 |
ENST00000062104.2
ENST00000346199.2
|
NNAT
|
neuronatin
|
|
chr3_+_10068095
Show fit
|
36.08 |
ENST00000287647.3
ENST00000383807.1
ENST00000383806.1
ENST00000419585.1
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr17_+_40834580
Show fit
|
32.81 |
ENST00000264638.4
|
CNTNAP1
|
contactin associated protein 1
|
|
chr12_-_6798523
Show fit
|
31.88 |
ENST00000319770.3
|
ZNF384
|
zinc finger protein 384
|
|
chr12_-_6798410
Show fit
|
31.62 |
ENST00000361959.3
ENST00000436774.2
ENST00000544482.1
|
ZNF384
|
zinc finger protein 384
|
|
chr3_+_187871060
Show fit
|
31.35 |
ENST00000448637.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr12_-_45270151
Show fit
|
30.82 |
ENST00000429094.2
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr12_-_45270077
Show fit
|
29.99 |
ENST00000551601.1
ENST00000549027.1
ENST00000452445.2
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr4_-_134070250
Show fit
|
26.28 |
ENST00000505289.1
ENST00000509715.1
|
RP11-9G1.3
|
RP11-9G1.3
|
|
chr14_+_24583836
Show fit
|
25.90 |
ENST00000559115.1
ENST00000558215.1
ENST00000557810.1
ENST00000561375.1
ENST00000446197.3
ENST00000559796.1
ENST00000560713.1
ENST00000560901.1
ENST00000559382.1
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr11_-_111784005
Show fit
|
24.43 |
ENST00000527899.1
|
CRYAB
|
crystallin, alpha B
|
|
chr14_-_24584138
Show fit
|
24.15 |
ENST00000558280.1
ENST00000561028.1
|
NRL
|
neural retina leucine zipper
|
|
chr11_-_111782696
Show fit
|
23.38 |
ENST00000227251.3
ENST00000526180.1
|
CRYAB
|
crystallin, alpha B
|
|
chr4_-_176733897
Show fit
|
22.15 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A
|
|
chr9_-_98079965
Show fit
|
21.76 |
ENST00000289081.3
|
FANCC
|
Fanconi anemia, complementation group C
|
|
chr6_-_29600832
Show fit
|
21.15 |
ENST00000377016.4
ENST00000376977.3
ENST00000377034.4
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr11_+_111783450
Show fit
|
21.14 |
ENST00000537382.1
|
HSPB2
|
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA.
|
|
chr11_+_111782934
Show fit
|
20.89 |
ENST00000304298.3
|
HSPB2
|
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA.
|
|
chr20_+_57430162
Show fit
|
20.89 |
ENST00000450130.1
ENST00000349036.3
ENST00000423897.1
|
GNAS
|
GNAS complex locus
|
|
chr2_+_25016282
Show fit
|
19.84 |
ENST00000260662.1
|
CENPO
|
centromere protein O
|
|
chr5_+_175792459
Show fit
|
19.21 |
ENST00000310389.5
|
ARL10
|
ADP-ribosylation factor-like 10
|
|
chr17_-_27278304
Show fit
|
18.70 |
ENST00000577226.1
|
PHF12
|
PHD finger protein 12
|
|
chr2_+_25015968
Show fit
|
18.68 |
ENST00000380834.2
ENST00000473706.1
|
CENPO
|
centromere protein O
|
|
chr11_-_111782484
Show fit
|
18.48 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B
|
|
chr6_+_163148973
Show fit
|
18.17 |
ENST00000366888.2
|
PACRG
|
PARK2 co-regulated
|
|
chrX_+_51928002
Show fit
|
17.40 |
ENST00000375626.3
|
MAGED4
|
melanoma antigen family D, 4
|
|
chr10_-_95360983
Show fit
|
16.97 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma
|
|
chr5_+_71403061
Show fit
|
16.45 |
ENST00000512974.1
ENST00000296755.7
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr11_-_12030629
Show fit
|
16.31 |
ENST00000396505.2
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3
|
|
chr14_+_101292445
Show fit
|
16.23 |
ENST00000429159.2
ENST00000520714.1
ENST00000522771.2
ENST00000424076.3
ENST00000423456.1
ENST00000521404.1
ENST00000556736.1
ENST00000451743.2
ENST00000398518.2
ENST00000554639.1
ENST00000452120.2
ENST00000519709.1
ENST00000412736.2
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr9_-_123476612
Show fit
|
16.17 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9
|
|
chr19_-_40919271
Show fit
|
14.91 |
ENST00000291825.7
ENST00000324001.7
|
PRX
|
periaxin
|
|
chr8_-_17555164
Show fit
|
14.66 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr17_-_7137582
Show fit
|
14.07 |
ENST00000575756.1
ENST00000575458.1
|
DVL2
|
dishevelled segment polarity protein 2
|
|
chr1_+_36690011
Show fit
|
14.02 |
ENST00000354618.5
ENST00000469141.2
ENST00000478853.1
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr1_-_38512450
Show fit
|
13.36 |
ENST00000373012.2
|
POU3F1
|
POU class 3 homeobox 1
|
|
chr5_+_161495038
Show fit
|
13.29 |
ENST00000393933.4
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2
|
|
chr12_+_57522258
Show fit
|
13.05 |
ENST00000553277.1
ENST00000243077.3
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chr5_+_161275320
Show fit
|
12.02 |
ENST00000437025.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr1_-_183387723
Show fit
|
11.95 |
ENST00000287713.6
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chrX_-_107979616
Show fit
|
11.67 |
ENST00000372129.2
|
IRS4
|
insulin receptor substrate 4
|
|
chr10_-_90712520
Show fit
|
11.63 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr7_+_119913688
Show fit
|
11.27 |
ENST00000331113.4
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2
|
|
chr9_-_86571628
Show fit
|
11.11 |
ENST00000376344.3
|
C9orf64
|
chromosome 9 open reading frame 64
|
|
chr2_-_51259292
Show fit
|
10.70 |
ENST00000401669.2
|
NRXN1
|
neurexin 1
|
|
chr6_+_31620191
Show fit
|
10.68 |
ENST00000375918.2
ENST00000375920.4
|
APOM
|
apolipoprotein M
|
|
chr1_+_226736446
Show fit
|
10.50 |
ENST00000366788.3
ENST00000366789.4
|
C1orf95
|
chromosome 1 open reading frame 95
|
|
chr11_+_45868957
Show fit
|
9.98 |
ENST00000443527.2
|
CRY2
|
cryptochrome 2 (photolyase-like)
|
|
chr14_+_29236269
Show fit
|
9.91 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1
|
|
chr2_-_51259229
Show fit
|
9.79 |
ENST00000405472.3
|
NRXN1
|
neurexin 1
|
|
chr15_-_81282133
Show fit
|
9.74 |
ENST00000261758.4
|
MESDC2
|
mesoderm development candidate 2
|
|
chr19_+_58790314
Show fit
|
9.54 |
ENST00000196548.5
ENST00000608843.1
|
ZNF8
ZNF8
|
Zinc finger protein 8
zinc finger protein 8
|
|
chr19_-_13617037
Show fit
|
9.41 |
ENST00000360228.5
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit
|
|
chr7_-_108096822
Show fit
|
9.34 |
ENST00000379028.3
ENST00000413765.2
ENST00000379022.4
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr5_-_168727786
Show fit
|
9.33 |
ENST00000332966.8
|
SLIT3
|
slit homolog 3 (Drosophila)
|
|
chrX_-_37706661
Show fit
|
9.30 |
ENST00000432389.2
ENST00000378581.3
|
DYNLT3
|
dynein, light chain, Tctex-type 3
|
|
chr7_-_108096765
Show fit
|
9.11 |
ENST00000379024.4
ENST00000351718.4
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr17_-_17875688
Show fit
|
9.02 |
ENST00000379504.3
ENST00000318094.10
ENST00000540946.1
ENST00000542206.1
ENST00000395739.4
ENST00000581396.1
ENST00000535933.1
ENST00000579586.1
|
TOM1L2
|
target of myb1-like 2 (chicken)
|
|
chr17_-_15244894
Show fit
|
8.97 |
ENST00000338696.2
ENST00000543896.1
ENST00000539245.1
ENST00000539316.1
ENST00000395930.1
|
TEKT3
|
tektin 3
|
|
chr1_-_45308616
Show fit
|
8.92 |
ENST00000447098.2
ENST00000372192.3
|
PTCH2
|
patched 2
|
|
chr10_+_99344104
Show fit
|
8.91 |
ENST00000555577.1
ENST00000370649.3
|
PI4K2A
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha
Phosphatidylinositol 4-kinase type 2-alpha; Uncharacterized protein
|
|
chr9_-_124989804
Show fit
|
8.87 |
ENST00000373755.2
ENST00000373754.2
|
LHX6
|
LIM homeobox 6
|
|
chr3_+_49449636
Show fit
|
8.86 |
ENST00000273590.3
|
TCTA
|
T-cell leukemia translocation altered
|
|
chr16_+_28834303
Show fit
|
8.75 |
ENST00000340394.8
ENST00000325215.6
ENST00000395547.2
ENST00000336783.4
ENST00000382686.4
ENST00000564304.1
|
ATXN2L
|
ataxin 2-like
|
|
chrX_-_140271249
Show fit
|
8.74 |
ENST00000370526.2
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr12_+_117176090
Show fit
|
8.45 |
ENST00000257575.4
ENST00000407967.3
ENST00000392549.2
|
RNFT2
|
ring finger protein, transmembrane 2
|
|
chr16_+_75256507
Show fit
|
8.39 |
ENST00000495583.1
|
CTRB1
|
chymotrypsinogen B1
|
|
chr5_+_161494770
Show fit
|
8.22 |
ENST00000414552.2
ENST00000361925.4
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2
|
|
chr9_-_123476719
Show fit
|
8.09 |
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9
|
|
chr9_-_122131696
Show fit
|
7.98 |
ENST00000373964.2
ENST00000265922.3
|
BRINP1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1
|
|
chr2_-_28113965
Show fit
|
7.69 |
ENST00000302188.3
|
RBKS
|
ribokinase
|
|
chr9_+_131218336
Show fit
|
7.65 |
ENST00000372814.3
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr2_-_51259641
Show fit
|
7.53 |
ENST00000406316.2
ENST00000405581.1
|
NRXN1
|
neurexin 1
|
|
chr17_+_17876127
Show fit
|
7.47 |
ENST00000582416.1
ENST00000313838.8
ENST00000411504.2
ENST00000581264.1
ENST00000399187.1
ENST00000479684.2
ENST00000584166.1
ENST00000585108.1
ENST00000399182.1
ENST00000579977.1
|
LRRC48
|
leucine rich repeat containing 48
|
|
chr21_-_34100244
Show fit
|
7.44 |
ENST00000382491.3
ENST00000357345.3
ENST00000429236.1
|
SYNJ1
|
synaptojanin 1
|
|
chr9_-_98279241
Show fit
|
7.43 |
ENST00000437951.1
ENST00000375274.2
ENST00000430669.2
ENST00000468211.2
|
PTCH1
|
patched 1
|
|
chr14_-_23834411
Show fit
|
7.25 |
ENST00000429593.2
|
EFS
|
embryonal Fyn-associated substrate
|
|
chr4_-_186697044
Show fit
|
7.25 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr3_-_73673991
Show fit
|
7.16 |
ENST00000308537.4
ENST00000263666.4
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr21_-_28217721
Show fit
|
7.13 |
ENST00000284984.3
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr2_-_51259528
Show fit
|
7.10 |
ENST00000404971.1
|
NRXN1
|
neurexin 1
|
|
chr1_-_111217603
Show fit
|
7.05 |
ENST00000369769.2
|
KCNA3
|
potassium voltage-gated channel, shaker-related subfamily, member 3
|
|
chr4_+_77870960
Show fit
|
6.96 |
ENST00000505788.1
ENST00000510515.1
ENST00000504637.1
|
SEPT11
|
septin 11
|
|
chr14_+_22984601
Show fit
|
6.88 |
ENST00000390509.1
|
TRAJ28
|
T cell receptor alpha joining 28
|
|
chr9_-_115818951
Show fit
|
6.85 |
ENST00000553380.1
ENST00000374227.3
|
ZFP37
|
ZFP37 zinc finger protein
|
|
chr21_-_34100341
Show fit
|
6.85 |
ENST00000382499.2
ENST00000433931.2
|
SYNJ1
|
synaptojanin 1
|
|
chr1_-_40105617
Show fit
|
6.84 |
ENST00000372852.3
|
HEYL
|
hes-related family bHLH transcription factor with YRPW motif-like
|
|
chr15_-_26108355
Show fit
|
6.81 |
ENST00000356865.6
|
ATP10A
|
ATPase, class V, type 10A
|
|
chr19_+_36208877
Show fit
|
6.67 |
ENST00000420124.1
ENST00000222270.7
ENST00000341701.1
|
KMT2B
|
Histone-lysine N-methyltransferase 2B
|
|
chr2_+_26915584
Show fit
|
6.67 |
ENST00000302909.3
|
KCNK3
|
potassium channel, subfamily K, member 3
|
|
chr19_+_42772659
Show fit
|
6.58 |
ENST00000572681.2
|
CIC
|
capicua transcriptional repressor
|
|
chr4_+_72052964
Show fit
|
6.58 |
ENST00000264485.5
ENST00000425175.1
|
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4
|
|
chr17_+_4843654
Show fit
|
6.39 |
ENST00000575111.1
|
RNF167
|
ring finger protein 167
|
|
chr2_-_68694390
Show fit
|
6.36 |
ENST00000377957.3
|
FBXO48
|
F-box protein 48
|
|
chr6_+_43739697
Show fit
|
6.25 |
ENST00000230480.6
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr3_+_4535155
Show fit
|
6.15 |
ENST00000544951.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1
|
|
chr14_+_24584508
Show fit
|
6.11 |
ENST00000559354.1
ENST00000560459.1
ENST00000559593.1
ENST00000396941.4
ENST00000396936.1
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr6_-_26027480
Show fit
|
5.98 |
ENST00000377364.3
|
HIST1H4B
|
histone cluster 1, H4b
|
|
chr5_-_137090028
Show fit
|
5.96 |
ENST00000314940.4
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr11_-_64527425
Show fit
|
5.80 |
ENST00000377432.3
|
PYGM
|
phosphorylase, glycogen, muscle
|
|
chr9_+_8858102
Show fit
|
5.78 |
ENST00000447950.1
ENST00000430766.1
|
RP11-75C9.1
|
RP11-75C9.1
|
|
chr2_-_208989225
Show fit
|
5.77 |
ENST00000264376.4
|
CRYGD
|
crystallin, gamma D
|
|
chrX_-_51812268
Show fit
|
5.76 |
ENST00000486010.1
ENST00000497164.1
ENST00000360134.6
ENST00000485287.1
ENST00000335504.5
ENST00000431659.1
|
MAGED4B
|
melanoma antigen family D, 4B
|
|
chrX_-_153236620
Show fit
|
5.74 |
ENST00000369984.4
|
HCFC1
|
host cell factor C1 (VP16-accessory protein)
|
|
chr1_-_182573514
Show fit
|
5.70 |
ENST00000367558.5
|
RGS16
|
regulator of G-protein signaling 16
|
|
chr4_+_62067860
Show fit
|
5.47 |
ENST00000514591.1
|
LPHN3
|
latrophilin 3
|
|
chrX_+_70364667
Show fit
|
5.33 |
ENST00000536169.1
ENST00000395855.2
ENST00000374051.3
ENST00000358741.3
|
NLGN3
|
neuroligin 3
|
|
chr9_-_115819039
Show fit
|
5.32 |
ENST00000555206.1
|
ZFP37
|
ZFP37 zinc finger protein
|
|
chr22_-_28197486
Show fit
|
5.23 |
ENST00000302326.4
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr7_-_4901625
Show fit
|
5.14 |
ENST00000404991.1
|
PAPOLB
|
poly(A) polymerase beta (testis specific)
|
|
chr7_-_92465868
Show fit
|
5.13 |
ENST00000424848.2
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr9_+_131219179
Show fit
|
5.09 |
ENST00000372791.3
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr3_-_33759541
Show fit
|
5.06 |
ENST00000468888.2
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr19_-_13617247
Show fit
|
5.03 |
ENST00000573710.2
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit
|
|
chr2_-_213403565
Show fit
|
5.00 |
ENST00000342788.4
ENST00000436443.1
|
ERBB4
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4
|
|
chr16_+_30675654
Show fit
|
4.96 |
ENST00000287468.5
ENST00000395073.2
|
FBRS
|
fibrosin
|
|
chr16_+_19183671
Show fit
|
4.95 |
ENST00000562711.2
|
SYT17
|
synaptotagmin XVII
|
|
chr12_-_4554780
Show fit
|
4.93 |
ENST00000228837.2
|
FGF6
|
fibroblast growth factor 6
|
|
chr15_-_78526855
Show fit
|
4.89 |
ENST00000541759.1
ENST00000558130.1
|
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1
|
|
chr1_+_164528866
Show fit
|
4.86 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr17_+_4843679
Show fit
|
4.85 |
ENST00000576229.1
|
RNF167
|
ring finger protein 167
|
|
chr11_+_33278811
Show fit
|
4.85 |
ENST00000303296.4
ENST00000379016.3
|
HIPK3
|
homeodomain interacting protein kinase 3
|
|
chr3_+_113616317
Show fit
|
4.84 |
ENST00000440446.2
ENST00000488680.1
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr11_+_10477733
Show fit
|
4.74 |
ENST00000528723.1
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr12_+_117176113
Show fit
|
4.68 |
ENST00000319176.7
|
RNFT2
|
ring finger protein, transmembrane 2
|
|
chr7_-_99679324
Show fit
|
4.67 |
ENST00000292393.5
ENST00000413658.2
ENST00000412947.1
ENST00000441298.1
ENST00000449785.1
ENST00000299667.4
ENST00000424697.1
|
ZNF3
|
zinc finger protein 3
|
|
chr4_-_186696425
Show fit
|
4.54 |
ENST00000430503.1
ENST00000319454.6
ENST00000450341.1
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr9_-_33264676
Show fit
|
4.51 |
ENST00000472232.3
ENST00000379704.2
|
BAG1
|
BCL2-associated athanogene
|
|
chr12_+_100661156
Show fit
|
4.39 |
ENST00000360820.2
|
SCYL2
|
SCY1-like 2 (S. cerevisiae)
|
|
chr13_+_20532807
Show fit
|
4.38 |
ENST00000382869.3
ENST00000382881.3
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr1_-_201081579
Show fit
|
4.36 |
ENST00000367338.3
ENST00000362061.3
|
CACNA1S
|
calcium channel, voltage-dependent, L type, alpha 1S subunit
|
|
chr17_+_7608511
Show fit
|
4.23 |
ENST00000226091.2
|
EFNB3
|
ephrin-B3
|
|
chr12_-_117175819
Show fit
|
4.18 |
ENST00000261318.3
ENST00000536380.1
|
C12orf49
|
chromosome 12 open reading frame 49
|
|
chr1_-_12677714
Show fit
|
4.11 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr15_-_78526942
Show fit
|
4.04 |
ENST00000258873.4
|
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1
|
|
chr17_-_40835076
Show fit
|
3.99 |
ENST00000591765.1
|
CCR10
|
chemokine (C-C motif) receptor 10
|
|
chr15_+_101142722
Show fit
|
3.92 |
ENST00000332783.7
ENST00000558747.1
ENST00000343276.4
|
ASB7
|
ankyrin repeat and SOCS box containing 7
|
|
chr1_-_112046289
Show fit
|
3.88 |
ENST00000241356.4
|
ADORA3
|
adenosine A3 receptor
|
|
chr17_-_76778339
Show fit
|
3.82 |
ENST00000591455.1
ENST00000446868.3
ENST00000361101.4
ENST00000589296.1
|
CYTH1
|
cytohesin 1
|
|
chr3_-_33759699
Show fit
|
3.78 |
ENST00000399362.4
ENST00000359576.5
ENST00000307312.7
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr1_-_156460391
Show fit
|
3.71 |
ENST00000360595.3
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr11_-_119991589
Show fit
|
3.71 |
ENST00000526881.1
|
TRIM29
|
tripartite motif containing 29
|
|
chr22_+_38035459
Show fit
|
3.48 |
ENST00000357436.4
|
SH3BP1
|
SH3-domain binding protein 1
|
|
chr1_-_21606013
Show fit
|
3.42 |
ENST00000357071.4
|
ECE1
|
endothelin converting enzyme 1
|
|
chr11_+_118478313
Show fit
|
3.41 |
ENST00000356063.5
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1
|
|
chr8_+_75736761
Show fit
|
3.33 |
ENST00000260113.2
|
PI15
|
peptidase inhibitor 15
|
|
chr19_+_4969116
Show fit
|
3.24 |
ENST00000588337.1
ENST00000159111.4
ENST00000381759.4
|
KDM4B
|
lysine (K)-specific demethylase 4B
|
|
chr17_+_7211280
Show fit
|
3.23 |
ENST00000419711.2
ENST00000571955.1
ENST00000573714.1
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr18_-_3880051
Show fit
|
3.23 |
ENST00000584874.1
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr9_+_34989638
Show fit
|
3.22 |
ENST00000453597.3
ENST00000335998.3
ENST00000312316.5
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr12_+_57853918
Show fit
|
3.22 |
ENST00000532291.1
ENST00000543426.1
ENST00000228682.2
ENST00000546141.1
|
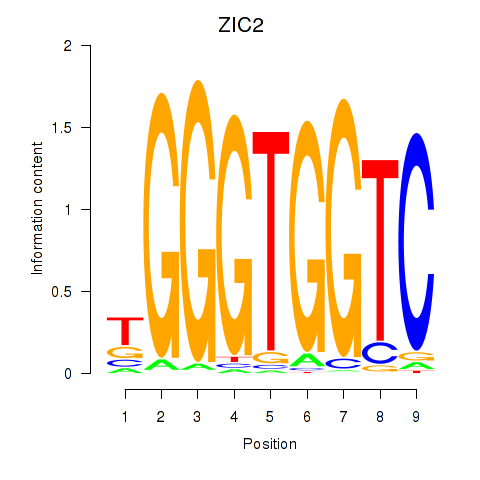
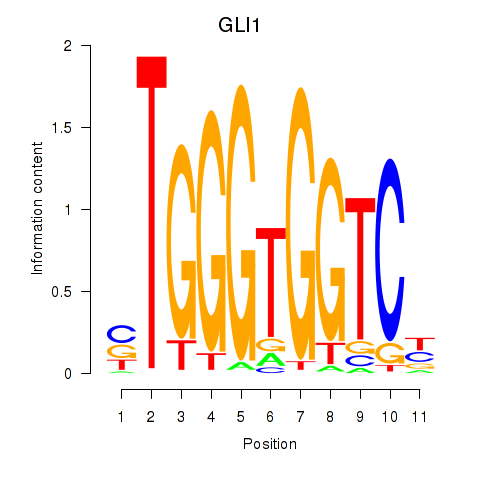
GLI1
|
GLI family zinc finger 1
|
|
chr9_-_33264557
Show fit
|
3.19 |
ENST00000473781.1
ENST00000488499.1
|
BAG1
|
BCL2-associated athanogene
|
|
chr1_-_112531777
Show fit
|
3.18 |
ENST00000315987.2
ENST00000302127.4
|
KCND3
|
potassium voltage-gated channel, Shal-related subfamily, member 3
|
|
chr19_+_45312347
Show fit
|
3.15 |
ENST00000270233.6
ENST00000591520.1
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group)
|
|
chr17_-_7137857
Show fit
|
3.15 |
ENST00000005340.5
|
DVL2
|
dishevelled segment polarity protein 2
|
|
chr9_+_131218408
Show fit
|
3.13 |
ENST00000351030.3
ENST00000604420.1
ENST00000535026.1
ENST00000448249.3
ENST00000393527.3
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr3_+_4535025
Show fit
|
3.10 |
ENST00000302640.8
ENST00000354582.6
ENST00000423119.2
ENST00000357086.4
ENST00000456211.2
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1
|
|
chr1_+_47264711
Show fit
|
3.09 |
ENST00000371923.4
ENST00000271153.4
ENST00000371919.4
|
CYP4B1
|
cytochrome P450, family 4, subfamily B, polypeptide 1
|
|
chr14_-_64971288
Show fit
|
3.08 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25
|
|
chrX_+_107334895
Show fit
|
3.07 |
ENST00000372232.3
ENST00000345734.3
ENST00000372254.3
|
ATG4A
|
autophagy related 4A, cysteine peptidase
|
|
chr13_+_20532848
Show fit
|
2.96 |
ENST00000382874.2
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr12_+_31477250
Show fit
|
2.92 |
ENST00000313737.4
|
AC024940.1
|
AC024940.1
|
|
chr6_-_31620403
Show fit
|
2.91 |
ENST00000451898.1
ENST00000439687.2
ENST00000362049.6
ENST00000424480.1
|
BAG6
|
BCL2-associated athanogene 6
|
|
chr5_+_52776449
Show fit
|
2.72 |
ENST00000396947.3
|
FST
|
follistatin
|
|
chr20_+_3451650
Show fit
|
2.66 |
ENST00000262919.5
|
ATRN
|
attractin
|
|
chr9_+_131218698
Show fit
|
2.65 |
ENST00000434106.3
ENST00000546203.1
ENST00000446274.1
ENST00000421776.2
ENST00000432065.2
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr17_+_17380294
Show fit
|
2.54 |
ENST00000268711.3
ENST00000580462.1
|
MED9
|
mediator complex subunit 9
|
|
chr3_+_6902794
Show fit
|
2.53 |
ENST00000357716.4
ENST00000486284.1
ENST00000389336.4
ENST00000403881.1
ENST00000402647.2
|
GRM7
|
glutamate receptor, metabotropic 7
|
|
chr20_-_50722183
Show fit
|
2.47 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein
|
|
chr20_+_3451732
Show fit
|
2.30 |
ENST00000446916.2
|
ATRN
|
attractin
|
|
chr16_-_68002456
Show fit
|
2.24 |
ENST00000576616.1
ENST00000572037.1
ENST00000338335.3
ENST00000422611.2
ENST00000316341.3
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporter), member 4
|
|
chr6_-_33160231
Show fit
|
2.24 |
ENST00000395194.1
ENST00000457788.1
ENST00000341947.2
ENST00000357486.1
ENST00000374714.1
ENST00000374713.1
ENST00000395197.1
ENST00000374712.1
ENST00000361917.1
ENST00000374708.4
|
COL11A2
|
collagen, type XI, alpha 2
|
|
chr13_+_20532900
Show fit
|
2.23 |
ENST00000382871.2
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr16_-_16317321
Show fit
|
2.14 |
ENST00000205557.7
ENST00000575728.1
|
ABCC6
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6
|
|
chr12_-_7848364
Show fit
|
2.13 |
ENST00000329913.3
|
GDF3
|
growth differentiation factor 3
|
|
chrX_-_30327495
Show fit
|
2.10 |
ENST00000453287.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1
|
|
chr6_-_31620455
Show fit
|
2.07 |
ENST00000437771.1
ENST00000404765.2
ENST00000375964.6
ENST00000211379.5
|
BAG6
|
BCL2-associated athanogene 6
|
|
chr17_-_50237343
Show fit
|
2.05 |
ENST00000575181.1
ENST00000570565.1
|
CA10
|
carbonic anhydrase X
|
|
chr6_-_43337180
Show fit
|
1.95 |
ENST00000318149.3
ENST00000361428.2
|
ZNF318
|
zinc finger protein 318
|
|
chr6_-_29595779
Show fit
|
1.93 |
ENST00000355973.3
ENST00000377012.4
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr3_+_170136642
Show fit
|
1.74 |
ENST00000064724.3
ENST00000486975.1
|
CLDN11
|
claudin 11
|
|
chr3_+_47844615
Show fit
|
1.65 |
ENST00000348968.4
|
DHX30
|
DEAH (Asp-Glu-Ala-His) box helicase 30
|
|
chr19_+_3708376
Show fit
|
1.64 |
ENST00000539908.2
|
TJP3
|
tight junction protein 3
|
|
chr17_-_50237374
Show fit
|
1.58 |
ENST00000442502.2
|
CA10
|
carbonic anhydrase X
|
|
chr17_+_73997419
Show fit
|
1.52 |
ENST00000425876.2
|
CDK3
|
cyclin-dependent kinase 3
|
|
chr19_-_51456321
Show fit
|
1.48 |
ENST00000391809.2
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr10_-_69455873
Show fit
|
1.44 |
ENST00000433211.2
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr8_-_41522779
Show fit
|
1.44 |
ENST00000522231.1
ENST00000314214.8
ENST00000348036.4
ENST00000457297.1
ENST00000522543.1
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr11_+_117049910
Show fit
|
1.27 |
ENST00000431081.2
ENST00000524842.1
|
SIDT2
|
SID1 transmembrane family, member 2
|
|
chr1_-_171621815
Show fit
|
1.21 |
ENST00000037502.6
|
MYOC
|
myocilin, trabecular meshwork inducible glucocorticoid response
|
|
chr16_+_86612112
Show fit
|
1.16 |
ENST00000320241.3
|
FOXL1
|
forkhead box L1
|
|
chr10_-_86001210
Show fit
|
1.16 |
ENST00000372105.3
|
LRIT1
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1
|
|
chr9_-_73483958
Show fit
|
1.13 |
ENST00000377101.1
ENST00000377106.1
ENST00000360823.2
ENST00000377105.1
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr14_-_88789581
Show fit
|
1.11 |
ENST00000319231.5
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr9_-_130639997
Show fit
|
1.04 |
ENST00000373176.1
|
AK1
|
adenylate kinase 1
|
|
chr3_+_137717571
Show fit
|
1.03 |
ENST00000343735.4
|
CLDN18
|
claudin 18
|