Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
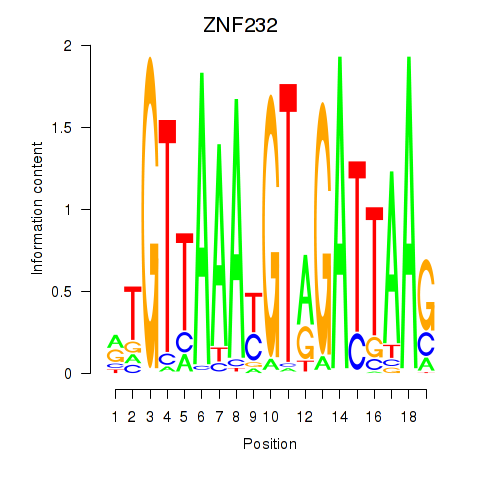
Results for ZNF232
Z-value: 0.61
Transcription factors associated with ZNF232
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF232
|
ENSG00000167840.9 | zinc finger protein 232 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF232 | hg19_v2_chr17_-_5015129_5015187 | -0.41 | 4.6e-10 | Click! |
Activity profile of ZNF232 motif
Sorted Z-values of ZNF232 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_88450612 | 33.23 |
ENST00000418378.1
ENST00000282470.6 |
SPARCL1
|
SPARC-like 1 (hevin) |
| chr3_+_127634069 | 23.62 |
ENST00000405109.1
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr15_-_60771128 | 20.93 |
ENST00000558512.1
ENST00000561114.1 |
NARG2
|
NMDA receptor regulated 2 |
| chr3_+_40566369 | 14.87 |
ENST00000403205.2
ENST00000310898.1 ENST00000339296.5 ENST00000431278.1 |
ZNF621
|
zinc finger protein 621 |
| chr7_+_142829162 | 9.67 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr14_-_21516590 | 8.21 |
ENST00000555026.1
|
NDRG2
|
NDRG family member 2 |
| chr3_-_185216811 | 8.02 |
ENST00000421852.1
|
TMEM41A
|
transmembrane protein 41A |
| chr17_-_6915646 | 6.71 |
ENST00000574377.1
ENST00000399541.2 ENST00000399540.2 ENST00000575727.1 ENST00000573939.1 |
AC027763.2
|
Uncharacterized protein |
| chr6_+_109416313 | 6.51 |
ENST00000521277.1
ENST00000517392.1 ENST00000407272.1 ENST00000336977.4 ENST00000519286.1 ENST00000518329.1 ENST00000522461.1 ENST00000518853.1 |
CEP57L1
|
centrosomal protein 57kDa-like 1 |
| chr16_-_4852915 | 6.45 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr9_-_115480303 | 6.33 |
ENST00000374234.1
ENST00000374238.1 ENST00000374236.1 ENST00000374242.4 |
INIP
|
INTS3 and NABP interacting protein |
| chr3_-_185216766 | 6.17 |
ENST00000296254.3
|
TMEM41A
|
transmembrane protein 41A |
| chr2_-_113522177 | 6.13 |
ENST00000541405.1
|
CKAP2L
|
cytoskeleton associated protein 2-like |
| chr8_-_143999259 | 5.43 |
ENST00000323110.2
|
CYP11B2
|
cytochrome P450, family 11, subfamily B, polypeptide 2 |
| chr6_-_46922659 | 5.30 |
ENST00000265417.7
|
GPR116
|
G protein-coupled receptor 116 |
| chrY_+_15016013 | 5.01 |
ENST00000360160.4
ENST00000454054.1 |
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr1_-_153029980 | 4.99 |
ENST00000392653.2
|
SPRR2A
|
small proline-rich protein 2A |
| chr3_+_8543561 | 4.84 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr5_+_177435986 | 4.83 |
ENST00000398106.2
|
FAM153C
|
family with sequence similarity 153, member C |
| chr3_+_8543533 | 4.69 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr4_+_96761238 | 4.53 |
ENST00000295266.4
|
PDHA2
|
pyruvate dehydrogenase (lipoamide) alpha 2 |
| chr1_-_79472365 | 4.51 |
ENST00000370742.3
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1 |
| chr20_+_4129426 | 4.40 |
ENST00000339123.6
ENST00000305958.4 ENST00000278795.3 |
SMOX
|
spermine oxidase |
| chr5_-_180076580 | 3.83 |
ENST00000502649.1
|
FLT4
|
fms-related tyrosine kinase 4 |
| chr2_-_113522248 | 3.77 |
ENST00000302450.6
|
CKAP2L
|
cytoskeleton associated protein 2-like |
| chr14_-_24711865 | 3.74 |
ENST00000399423.4
ENST00000267415.7 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr14_-_24711806 | 3.74 |
ENST00000540705.1
ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chrX_+_100805496 | 3.73 |
ENST00000372829.3
|
ARMCX1
|
armadillo repeat containing, X-linked 1 |
| chrX_-_130423386 | 3.51 |
ENST00000370903.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr18_+_61637159 | 3.45 |
ENST00000397985.2
ENST00000353706.2 ENST00000542677.1 ENST00000397988.3 ENST00000448851.1 |
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
| chr10_-_27389392 | 3.37 |
ENST00000376087.4
|
ANKRD26
|
ankyrin repeat domain 26 |
| chr11_+_66276550 | 3.18 |
ENST00000419755.3
|
CTD-3074O7.11
|
Bardet-Biedl syndrome 1 protein |
| chr9_+_5231413 | 2.61 |
ENST00000239316.4
|
INSL4
|
insulin-like 4 (placenta) |
| chr7_-_83278322 | 2.59 |
ENST00000307792.3
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr10_-_115904361 | 2.42 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr15_-_79383102 | 2.39 |
ENST00000558480.2
ENST00000419573.3 |
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1 |
| chr10_-_61900762 | 2.37 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr1_+_182419261 | 2.28 |
ENST00000294854.8
ENST00000542961.1 |
RGSL1
|
regulator of G-protein signaling like 1 |
| chr17_-_3301704 | 2.21 |
ENST00000322608.2
|
OR1E1
|
olfactory receptor, family 1, subfamily E, member 1 |
| chr17_+_26800648 | 2.05 |
ENST00000545060.1
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr1_-_204654826 | 1.92 |
ENST00000367177.3
|
LRRN2
|
leucine rich repeat neuronal 2 |
| chr2_+_241807870 | 1.91 |
ENST00000307503.3
|
AGXT
|
alanine-glyoxylate aminotransferase |
| chr14_-_107131560 | 1.82 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66 |
| chr9_+_117085336 | 1.77 |
ENST00000259396.8
ENST00000538816.1 |
ORM1
|
orosomucoid 1 |
| chr17_+_26800296 | 1.72 |
ENST00000444914.3
ENST00000314669.5 |
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr15_-_74374891 | 1.68 |
ENST00000290438.3
|
GOLGA6A
|
golgin A6 family, member A |
| chr17_+_26800756 | 1.66 |
ENST00000537681.1
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr15_-_56757329 | 1.64 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr16_+_30662184 | 1.58 |
ENST00000300835.4
|
PRR14
|
proline rich 14 |
| chr1_-_160549235 | 1.56 |
ENST00000368054.3
ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84
|
CD84 molecule |
| chr13_+_27825706 | 1.52 |
ENST00000272274.4
ENST00000319826.4 ENST00000326092.4 |
RPL21
|
ribosomal protein L21 |
| chr2_-_31637560 | 1.29 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chr2_+_234826016 | 1.22 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr5_-_180076613 | 1.16 |
ENST00000261937.6
ENST00000393347.3 |
FLT4
|
fms-related tyrosine kinase 4 |
| chr16_-_20367584 | 1.05 |
ENST00000570689.1
|
UMOD
|
uromodulin |
| chr21_+_38792602 | 0.98 |
ENST00000398960.2
ENST00000398956.2 |
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chr16_+_30662360 | 0.86 |
ENST00000542965.2
|
PRR14
|
proline rich 14 |
| chrY_+_15016725 | 0.84 |
ENST00000336079.3
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chrX_-_100548045 | 0.76 |
ENST00000372907.3
ENST00000372905.2 |
TAF7L
|
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa |
| chr10_+_50817141 | 0.75 |
ENST00000339797.1
|
CHAT
|
choline O-acetyltransferase |
| chr2_-_74618964 | 0.56 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr1_+_174669653 | 0.44 |
ENST00000325589.5
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr3_+_8543393 | 0.33 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr14_-_106816253 | 0.31 |
ENST00000390615.2
|
IGHV3-33
|
immunoglobulin heavy variable 3-33 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF232
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 7.5 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.7 | 4.4 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.6 | 1.9 | GO:0006522 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.6 | 2.4 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.5 | 5.4 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.5 | 8.2 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.4 | 1.3 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.4 | 5.0 | GO:0060312 | regulation of blood vessel remodeling(GO:0060312) |
| 0.4 | 9.9 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.4 | 9.7 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.3 | 1.6 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.3 | 4.5 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.2 | 1.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.2 | 1.8 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.2 | 1.0 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.2 | 3.4 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 2.4 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 1.6 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 5.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 5.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 2.6 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 6.5 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 1.0 | GO:0090312 | positive regulation of protein deacetylation(GO:0090312) |
| 0.0 | 2.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 5.4 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.0 | 6.5 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 19.5 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 0.8 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 6.3 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.0 | 17.2 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.8 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 2.6 | GO:0007565 | female pregnancy(GO:0007565) |
| 0.0 | 1.5 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 1.5 | 7.5 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.5 | 4.5 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.3 | 23.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 2.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 9.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 5.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 32.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 10.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 9.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.9 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 11.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.8 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 5.8 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 10.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 12.0 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 9.9 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 1.2 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 5.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) organelle inner membrane(GO:0019866) |
| 0.0 | 1.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.8 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 5.0 | GO:0043235 | receptor complex(GO:0043235) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 1.8 | 5.4 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 1.5 | 4.4 | GO:0052895 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 1.1 | 4.5 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.6 | 10.7 | GO:0019864 | IgG binding(GO:0019864) |
| 0.4 | 33.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.4 | 3.5 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.4 | 5.0 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 6.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 0.8 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.2 | 2.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 1.3 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 7.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 1.9 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 5.8 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 2.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 1.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 2.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 2.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 17.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.4 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 3.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 9.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 14.9 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 33.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 8.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 7.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 2.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 5.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 5.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 4.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 7.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 2.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 2.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 14.9 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.8 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |