Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
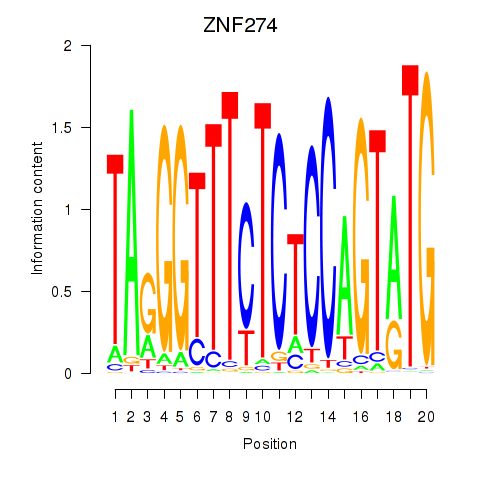
Results for ZNF274
Z-value: 0.62
Transcription factors associated with ZNF274
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF274
|
ENSG00000171606.13 | zinc finger protein 274 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF274 | hg19_v2_chr19_+_58694396_58694485 | 0.40 | 1.0e-09 | Click! |
Activity profile of ZNF274 motif
Sorted Z-values of ZNF274 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_119737806 | 18.15 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr5_+_32531893 | 16.05 |
ENST00000512913.1
|
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr18_+_9103957 | 15.47 |
ENST00000400033.1
|
NDUFV2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| chr12_-_54653313 | 14.24 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr1_+_45205498 | 13.68 |
ENST00000372218.4
|
KIF2C
|
kinesin family member 2C |
| chr11_+_16760161 | 13.51 |
ENST00000524439.1
ENST00000422258.2 ENST00000528634.1 ENST00000525684.1 |
C11orf58
|
chromosome 11 open reading frame 58 |
| chr1_+_45205478 | 13.17 |
ENST00000452259.1
ENST00000372224.4 |
KIF2C
|
kinesin family member 2C |
| chr16_+_29819096 | 11.69 |
ENST00000568411.1
ENST00000563012.1 ENST00000562557.1 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr2_-_96971259 | 10.72 |
ENST00000349783.5
|
SNRNP200
|
small nuclear ribonucleoprotein 200kDa (U5) |
| chr12_+_113344755 | 10.71 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr22_-_43036607 | 10.70 |
ENST00000505920.1
|
ATP5L2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2 |
| chr6_+_30687978 | 9.94 |
ENST00000327892.8
ENST00000435534.1 |
TUBB
|
tubulin, beta class I |
| chr16_+_29819372 | 9.01 |
ENST00000568544.1
ENST00000569978.1 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr16_+_29818857 | 8.46 |
ENST00000567444.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr16_+_29819446 | 8.29 |
ENST00000568282.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr10_+_5488564 | 4.65 |
ENST00000449083.1
ENST00000380359.3 |
NET1
|
neuroepithelial cell transforming 1 |
| chr6_+_31553901 | 4.04 |
ENST00000418507.2
ENST00000438075.2 ENST00000376100.3 ENST00000376111.4 |
LST1
|
leukocyte specific transcript 1 |
| chr17_-_7145106 | 4.03 |
ENST00000577035.1
|
GABARAP
|
GABA(A) receptor-associated protein |
| chr3_+_37903432 | 3.66 |
ENST00000443503.2
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr17_-_41132410 | 3.28 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr3_+_50284321 | 2.98 |
ENST00000451956.1
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr1_-_114414316 | 2.98 |
ENST00000528414.1
ENST00000538253.1 ENST00000460620.1 ENST00000420377.2 ENST00000525799.1 ENST00000359785.5 |
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr6_+_31553978 | 2.73 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr22_+_39353527 | 2.68 |
ENST00000249116.2
|
APOBEC3A
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A |
| chr4_+_15704679 | 2.23 |
ENST00000382346.3
|
BST1
|
bone marrow stromal cell antigen 1 |
| chr4_-_121993673 | 1.89 |
ENST00000379692.4
|
NDNF
|
neuron-derived neurotrophic factor |
| chr19_+_40195101 | 1.83 |
ENST00000360675.3
ENST00000601802.1 |
LGALS14
|
lectin, galactoside-binding, soluble, 14 |
| chr12_+_50135327 | 1.82 |
ENST00000549966.1
ENST00000547832.1 ENST00000547187.1 ENST00000548894.1 ENST00000546914.1 ENST00000552699.1 ENST00000267115.5 |
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr19_-_19774473 | 1.56 |
ENST00000357324.6
|
ATP13A1
|
ATPase type 13A1 |
| chr20_+_60174827 | 1.41 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr2_+_11864458 | 1.20 |
ENST00000396098.1
ENST00000396099.1 ENST00000425416.2 |
LPIN1
|
lipin 1 |
| chr2_-_89292422 | 1.05 |
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8 |
| chr19_+_6887571 | 0.96 |
ENST00000250572.8
ENST00000381407.5 ENST00000312053.4 ENST00000450315.3 ENST00000381404.4 |
EMR1
|
egf-like module containing, mucin-like, hormone receptor-like 1 |
| chr4_+_15704573 | 0.77 |
ENST00000265016.4
|
BST1
|
bone marrow stromal cell antigen 1 |
| chr6_-_32784687 | 0.46 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF274
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 18.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 3.6 | 10.7 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 2.7 | 26.9 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.0 | 3.0 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.4 | 2.7 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.4 | 16.0 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.4 | 37.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.4 | 4.7 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.3 | 1.8 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.3 | 10.7 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 4.0 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 15.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 3.0 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.2 | 1.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.2 | 1.9 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.2 | 1.6 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.2 | 0.5 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 9.9 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.1 | 10.7 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.1 | 3.0 | GO:2001044 | regulation of integrin-mediated signaling pathway(GO:2001044) |
| 0.1 | 6.8 | GO:0050672 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 0.0 | 0.4 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 14.2 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 1.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 3.7 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 15.5 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 1.0 | 14.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.8 | 26.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.7 | 9.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.6 | 10.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.4 | 10.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 18.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 4.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 3.0 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 3.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 11.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 2.7 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 26.9 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 1.1 | 10.7 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 1.0 | 3.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.7 | 15.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.5 | 2.7 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.5 | 9.9 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.4 | 1.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.4 | 10.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.3 | 16.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.3 | 37.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.3 | 1.8 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.3 | 4.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 18.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.2 | 14.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.2 | 4.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 1.6 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 10.7 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 3.0 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.5 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 6.6 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 41.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 3.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 3.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 4.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 26.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 10.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.2 | 15.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 3.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 10.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 14.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 9.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 4.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |