Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for ZNF524
Z-value: 1.03
Transcription factors associated with ZNF524
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF524
|
ENSG00000171443.6 | zinc finger protein 524 |
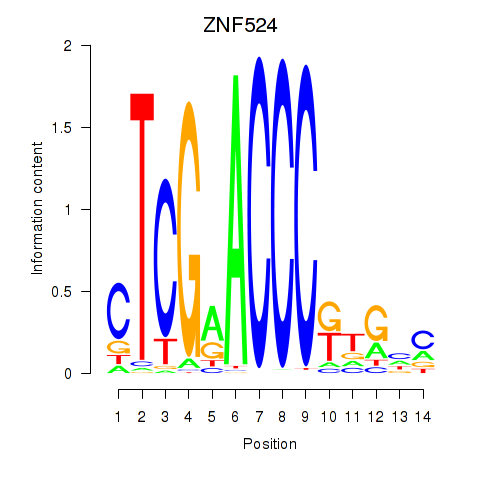
Activity profile of ZNF524 motif
Sorted Z-values of ZNF524 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF524
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.5 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 3.5 | 14.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 3.1 | 9.2 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 2.4 | 9.6 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 2.4 | 7.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 2.3 | 9.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 2.1 | 6.3 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 1.8 | 7.1 | GO:1902612 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 1.8 | 8.8 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 1.7 | 5.2 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 1.6 | 4.9 | GO:0030451 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 1.6 | 4.9 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 1.6 | 4.7 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 1.6 | 4.7 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 1.6 | 4.7 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 1.5 | 6.2 | GO:0014028 | notochord formation(GO:0014028) |
| 1.5 | 3.0 | GO:2001176 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 1.5 | 12.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 1.5 | 10.6 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 1.5 | 4.4 | GO:2000547 | dendritic cell dendrite assembly(GO:0097026) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 1.4 | 5.7 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 1.3 | 7.8 | GO:0051012 | microtubule sliding(GO:0051012) |
| 1.3 | 5.2 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 1.3 | 3.8 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 1.3 | 5.0 | GO:0000023 | maltose metabolic process(GO:0000023) diaphragm contraction(GO:0002086) |
| 1.2 | 3.7 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 1.2 | 3.7 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 1.2 | 7.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 1.2 | 5.9 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 1.1 | 3.4 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 1.1 | 6.7 | GO:0030421 | defecation(GO:0030421) |
| 1.1 | 5.5 | GO:0014055 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) |
| 1.1 | 3.3 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) establishment of meiotic spindle localization(GO:0051295) |
| 1.1 | 3.3 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 1.1 | 6.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 1.1 | 4.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 1.1 | 3.2 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 1.0 | 4.2 | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) |
| 1.0 | 4.1 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 1.0 | 7.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 1.0 | 4.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 1.0 | 1.9 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 1.0 | 10.6 | GO:0002775 | antimicrobial peptide production(GO:0002775) antibacterial peptide production(GO:0002778) |
| 0.9 | 1.9 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.9 | 2.8 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.9 | 3.7 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.9 | 1.8 | GO:0007605 | sensory perception of sound(GO:0007605) sensory perception of mechanical stimulus(GO:0050954) |
| 0.9 | 3.7 | GO:0035720 | signal transduction downstream of smoothened(GO:0007227) intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.9 | 2.7 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.9 | 3.6 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.9 | 2.7 | GO:0071988 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.9 | 4.4 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) |
| 0.9 | 1.8 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.9 | 4.4 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.9 | 3.5 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.9 | 6.0 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.9 | 2.6 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.8 | 14.4 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.8 | 6.7 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.8 | 4.1 | GO:0051946 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.8 | 3.2 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.8 | 2.4 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.8 | 2.4 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.8 | 2.4 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.8 | 2.4 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.8 | 3.1 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.8 | 3.9 | GO:0009440 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.8 | 2.3 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) |
| 0.8 | 2.3 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.8 | 2.3 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.8 | 2.3 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.7 | 7.4 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.7 | 2.9 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.7 | 2.9 | GO:1904640 | response to methionine(GO:1904640) |
| 0.7 | 2.8 | GO:0072255 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.7 | 6.4 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.7 | 2.1 | GO:0048936 | neurofilament bundle assembly(GO:0033693) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.7 | 0.7 | GO:2000864 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.7 | 1.4 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.7 | 7.6 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.7 | 2.1 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.7 | 2.7 | GO:0042418 | epinephrine metabolic process(GO:0042414) epinephrine biosynthetic process(GO:0042418) |
| 0.7 | 4.7 | GO:0006489 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.7 | 2.0 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.7 | 3.9 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.7 | 2.0 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.7 | 6.5 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.6 | 1.9 | GO:1904970 | intermicrovillar adhesion(GO:0090675) brush border assembly(GO:1904970) |
| 0.6 | 3.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.6 | 2.5 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.6 | 2.5 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.6 | 1.9 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.6 | 1.8 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) frontal suture morphogenesis(GO:0060364) |
| 0.6 | 3.0 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.6 | 2.3 | GO:0072366 | regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.6 | 2.3 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.6 | 2.3 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.6 | 1.7 | GO:0070781 | response to biotin(GO:0070781) |
| 0.6 | 2.2 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.6 | 4.4 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.5 | 1.1 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.5 | 1.6 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.5 | 3.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.5 | 1.6 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.5 | 2.1 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.5 | 2.6 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.5 | 1.0 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.5 | 2.0 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.5 | 6.0 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.5 | 1.5 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.5 | 2.9 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.5 | 4.9 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.5 | 2.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.5 | 3.8 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.5 | 1.4 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.5 | 3.8 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.5 | 2.4 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.5 | 2.8 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.5 | 6.0 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.5 | 2.3 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.5 | 0.9 | GO:0022028 | tangential migration from the subventricular zone to the olfactory bulb(GO:0022028) |
| 0.4 | 5.4 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.4 | 1.8 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.4 | 1.3 | GO:0090298 | negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.4 | 0.9 | GO:0060374 | erythropoietin-mediated signaling pathway(GO:0038162) mast cell differentiation(GO:0060374) |
| 0.4 | 2.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.4 | 2.6 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.4 | 1.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.4 | 1.7 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.4 | 6.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.4 | 4.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.4 | 2.9 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.4 | 2.5 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.4 | 1.2 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.4 | 2.0 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.4 | 2.0 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.4 | 0.8 | GO:0071623 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.4 | 3.6 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.4 | 0.8 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.4 | 1.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.4 | 3.0 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.4 | 3.4 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.4 | 7.2 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.4 | 1.9 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.4 | 2.6 | GO:0090045 | positive regulation of deacetylase activity(GO:0090045) positive regulation of histone deacetylase activity(GO:1901727) |
| 0.4 | 2.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.4 | 2.9 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.4 | 0.7 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.4 | 1.4 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.4 | 2.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.3 | 0.7 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.3 | 3.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.3 | 1.7 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.3 | 3.8 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.3 | 4.4 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.3 | 1.3 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 0.3 | 2.0 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.3 | 2.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.3 | 1.7 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.3 | 2.0 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.3 | 3.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.3 | 1.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.3 | 2.0 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.3 | 4.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.3 | 4.9 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.3 | 1.3 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.3 | 6.8 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.3 | 2.2 | GO:0007292 | female gamete generation(GO:0007292) |
| 0.3 | 1.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.3 | 1.0 | GO:0098759 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.3 | 1.6 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.3 | 2.5 | GO:0042148 | strand invasion(GO:0042148) |
| 0.3 | 7.9 | GO:0035909 | aorta morphogenesis(GO:0035909) |
| 0.3 | 0.3 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.3 | 4.4 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.3 | 0.9 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.3 | 1.2 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.3 | 0.9 | GO:1903935 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.3 | 6.9 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.3 | 1.8 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.3 | 1.4 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.3 | 4.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 0.8 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.3 | 0.8 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.3 | 2.8 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.3 | 1.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.3 | 0.3 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.3 | 1.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.3 | 1.1 | GO:0007270 | neuron-neuron synaptic transmission(GO:0007270) |
| 0.3 | 0.8 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 0.3 | 1.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.3 | 1.9 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.3 | 5.6 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.3 | 1.9 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.3 | 1.9 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.3 | 1.8 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.3 | 2.4 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.3 | 4.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.3 | 3.6 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.3 | 2.3 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.3 | 1.0 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.3 | 1.5 | GO:1904627 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.3 | 1.0 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.3 | 2.5 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.2 | 3.0 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 0.7 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.2 | 1.2 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.2 | 3.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 1.4 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.2 | 1.4 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 0.2 | 0.9 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.2 | 2.6 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.2 | 0.7 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.2 | 4.2 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.2 | 1.2 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.2 | 9.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 0.7 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.2 | 2.5 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.2 | 1.8 | GO:2000667 | positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.2 | 0.7 | GO:0035283 | rhombomere 3 development(GO:0021569) rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.2 | 0.9 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.2 | 2.5 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.2 | 0.7 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.2 | 1.7 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.2 | 1.3 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.2 | 0.8 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.2 | 1.0 | GO:0097191 | extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.2 | 2.2 | GO:0050672 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) negative regulation of leukocyte proliferation(GO:0070664) |
| 0.2 | 3.2 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.2 | 11.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 2.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.2 | 1.4 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 2.4 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.2 | 4.8 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.2 | 1.2 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.2 | 2.0 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.2 | 2.2 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.2 | 2.6 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.2 | 1.6 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.2 | 2.0 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.2 | 1.7 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.2 | 2.5 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.2 | 4.7 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.2 | 4.1 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.2 | 3.4 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.2 | 1.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.2 | 1.5 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.2 | 4.6 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.2 | 0.7 | GO:0060849 | radial pattern formation(GO:0009956) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.2 | 1.3 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.2 | 2.0 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.2 | 3.9 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 4.3 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.2 | 1.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 5.3 | GO:0051567 | histone H3-K9 methylation(GO:0051567) |
| 0.2 | 1.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.2 | 0.5 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.2 | 3.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.2 | 1.7 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 1.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 1.0 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.2 | 3.3 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.2 | 1.2 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.2 | 3.6 | GO:0036499 | PERK-mediated unfolded protein response(GO:0036499) |
| 0.2 | 0.5 | GO:1903939 | regulation of TORC2 signaling(GO:1903939) |
| 0.2 | 5.9 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.2 | 1.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 1.5 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 6.2 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.2 | 2.1 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.2 | 4.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 3.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 2.9 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.2 | 1.7 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.2 | 0.6 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 | 6.1 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.2 | 5.1 | GO:1903861 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.2 | 2.3 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.2 | 0.8 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 1.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 2.1 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 0.3 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 2.3 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.1 | 0.7 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.1 | 3.0 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.1 | 2.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.8 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.1 | 1.0 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.1 | 1.9 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 3.2 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.1 | 5.3 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.1 | 1.6 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.7 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.7 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.4 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.1 | 2.4 | GO:0001976 | neurological system process involved in regulation of systemic arterial blood pressure(GO:0001976) |
| 0.1 | 1.2 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.1 | 1.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 2.5 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 4.5 | GO:0007140 | male meiosis(GO:0007140) |
| 0.1 | 1.0 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 3.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.4 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 1.8 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 4.1 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.1 | 0.5 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 6.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.5 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 2.2 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.5 | GO:0051971 | positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.1 | 1.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 3.2 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 1.2 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.9 | GO:0098877 | neurotransmitter receptor transport to plasma membrane(GO:0098877) neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) neurotransmitter receptor transport to postsynaptic membrane(GO:0098969) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) establishment of protein localization to postsynaptic membrane(GO:1903540) |
| 0.1 | 1.5 | GO:0061436 | regulation of water loss via skin(GO:0033561) establishment of skin barrier(GO:0061436) |
| 0.1 | 2.4 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 0.8 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.9 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.2 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.1 | 1.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 1.7 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 1.4 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.1 | 2.0 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 0.9 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.1 | 1.3 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.1 | 1.7 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 2.0 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 2.4 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 1.6 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 2.6 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 1.8 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 1.7 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.1 | 1.0 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.3 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 5.3 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 0.6 | GO:0036100 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.1 | 1.8 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.1 | 0.4 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.1 | 1.8 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 1.4 | GO:0097502 | mannosylation(GO:0097502) |
| 0.1 | 0.5 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 1.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.6 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.4 | GO:2001171 | positive regulation of ATP biosynthetic process(GO:2001171) |
| 0.1 | 0.4 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.1 | 0.4 | GO:0045007 | depurination(GO:0045007) |
| 0.1 | 1.0 | GO:0050918 | positive chemotaxis(GO:0050918) |
| 0.1 | 2.2 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.8 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 1.7 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 1.1 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.1 | 3.4 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 2.7 | GO:0070873 | regulation of glycogen metabolic process(GO:0070873) |
| 0.1 | 0.2 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.1 | 0.6 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.2 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.1 | 1.5 | GO:0046135 | pyrimidine nucleoside catabolic process(GO:0046135) |
| 0.1 | 3.4 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 1.6 | GO:0048265 | response to pain(GO:0048265) |
| 0.1 | 0.2 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.1 | 0.6 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 2.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 1.4 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.1 | 1.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 2.0 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.7 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 2.3 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 1.6 | GO:0055010 | ventricular cardiac muscle tissue morphogenesis(GO:0055010) |
| 0.1 | 0.7 | GO:0009615 | response to virus(GO:0009615) |
| 0.1 | 1.0 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.1 | 0.4 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 0.7 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 1.7 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 4.8 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 7.6 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 2.0 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.1 | 1.5 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 1.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.2 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.6 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.2 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.1 | 0.5 | GO:0009452 | 7-methylguanosine mRNA capping(GO:0006370) 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.1 | 1.7 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 | 0.4 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 1.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 4.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.3 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 1.4 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 0.9 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 4.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 1.5 | GO:0001889 | liver development(GO:0001889) hepaticobiliary system development(GO:0061008) |
| 0.1 | 3.6 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 0.8 | GO:0045187 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 1.1 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 0.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 0.9 | GO:0007129 | synapsis(GO:0007129) |
| 0.1 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 1.1 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.2 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 2.8 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 0.5 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 1.5 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 2.5 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 1.1 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.7 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 1.2 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.4 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.4 | GO:0034138 | toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.0 | 1.7 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 2.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.4 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 1.2 | GO:0035924 | cellular response to vascular endothelial growth factor stimulus(GO:0035924) |
| 0.0 | 0.7 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.3 | GO:0021894 | cerebral cortex GABAergic interneuron differentiation(GO:0021892) cerebral cortex GABAergic interneuron development(GO:0021894) GABAergic neuron differentiation(GO:0097154) |
| 0.0 | 1.1 | GO:1904357 | negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.0 | 0.3 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 5.2 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.3 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.5 | GO:0010720 | positive regulation of cell development(GO:0010720) positive regulation of neurogenesis(GO:0050769) |
| 0.0 | 0.2 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 1.1 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 1.2 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.3 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 1.1 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.8 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.3 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 1.7 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 2.3 | GO:0050890 | cognition(GO:0050890) |
| 0.0 | 1.9 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.9 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.4 | GO:1901216 | positive regulation of neuron death(GO:1901216) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 1.1 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 1.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.5 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 1.1 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.4 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.3 | GO:0000732 | strand displacement(GO:0000732) |
| 0.0 | 0.3 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.9 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 0.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 2.9 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 1.1 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 5.4 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 1.3 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.0 | 1.0 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.0 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.0 | 2.2 | GO:0042445 | hormone metabolic process(GO:0042445) |
| 0.0 | 0.9 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.0 | 0.4 | GO:0018210 | peptidyl-threonine phosphorylation(GO:0018107) peptidyl-threonine modification(GO:0018210) |
| 0.0 | 0.3 | GO:0072600 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.4 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.4 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.0 | GO:0000187 | activation of MAPK activity(GO:0000187) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.2 | GO:1990745 | EARP complex(GO:1990745) |
| 2.2 | 17.7 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 1.7 | 1.7 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 1.7 | 6.7 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 1.6 | 6.5 | GO:0045160 | myosin I complex(GO:0045160) |
| 1.2 | 3.6 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 1.2 | 7.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 1.1 | 6.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 1.0 | 4.8 | GO:0001652 | granular component(GO:0001652) |
| 0.9 | 8.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.9 | 2.7 | GO:0061673 | cortical microtubule(GO:0055028) mitotic spindle astral microtubule(GO:0061673) |
| 0.9 | 3.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.8 | 3.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.8 | 5.4 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.7 | 2.9 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.7 | 4.4 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.6 | 7.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.6 | 3.8 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.6 | 3.7 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.6 | 2.4 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.6 | 2.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.6 | 7.9 | GO:0042599 | lamellar body(GO:0042599) |
| 0.6 | 2.8 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.5 | 3.2 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.5 | 1.5 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.5 | 4.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.5 | 5.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.5 | 3.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.5 | 8.2 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.5 | 1.9 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.5 | 2.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.4 | 2.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.4 | 2.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.4 | 1.3 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.4 | 2.5 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.4 | 1.9 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.4 | 3.8 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.4 | 3.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.4 | 1.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.4 | 2.2 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.4 | 11.9 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.4 | 2.5 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 4.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.3 | 1.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.3 | 1.6 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.3 | 1.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.3 | 2.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.3 | 0.9 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.3 | 4.5 | GO:0005883 | neurofilament(GO:0005883) |
| 0.3 | 2.4 | GO:0071204 | U7 snRNP(GO:0005683) histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.3 | 2.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.3 | 0.9 | GO:0098984 | symmetric synapse(GO:0032280) neuron to neuron synapse(GO:0098984) |
| 0.3 | 1.7 | GO:0042825 | TAP complex(GO:0042825) |
| 0.3 | 2.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.3 | 1.9 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.3 | 1.3 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.2 | 2.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.2 | 5.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 3.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 2.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 0.9 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 2.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 3.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.2 | 2.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 0.8 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.2 | 17.6 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.2 | 4.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.2 | 1.7 | GO:0032009 | early phagosome(GO:0032009) |
| 0.2 | 1.9 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.2 | 5.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 3.4 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.2 | 20.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.2 | 2.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 2.4 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 0.7 | GO:0043259 | laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.2 | 5.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 8.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 2.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 0.9 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.2 | 1.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.2 | 2.0 | GO:0010369 | chromocenter(GO:0010369) |
| 0.2 | 5.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 8.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.4 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.1 | 1.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 3.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 2.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 8.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 3.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 5.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 3.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.4 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 1.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 1.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 2.1 | GO:0098553 | integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 1.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.2 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.1 | 2.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 3.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.5 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 2.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 0.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.9 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 0.9 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.1 | 3.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.8 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 6.5 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.9 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 1.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.7 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 2.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 14.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 5.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 1.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.8 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 5.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 36.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 7.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 12.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 4.2 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 0.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 18.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 2.0 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 3.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 1.7 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 7.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 1.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 5.1 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.1 | 0.6 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.5 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.1 | 1.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 2.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 2.7 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 1.1 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.8 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 60.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.7 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 2.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 4.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 9.7 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.7 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 1.6 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.5 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.8 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 1.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.7 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.6 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 1.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.7 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 1.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.3 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 15.9 | GO:0097001 | ceramide binding(GO:0097001) |
| 3.5 | 14.1 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 3.0 | 9.0 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 2.6 | 10.6 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 2.5 | 7.4 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 2.1 | 6.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 1.8 | 14.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 1.7 | 5.0 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 1.6 | 4.9 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 1.6 | 6.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 1.5 | 23.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 1.4 | 7.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 1.3 | 8.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 1.3 | 5.2 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 1.3 | 3.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 1.2 | 3.7 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 1.2 | 7.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 1.2 | 4.7 | GO:0047708 | biotinidase activity(GO:0047708) |
| 1.1 | 4.5 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 1.1 | 11.2 | GO:0009374 | biotin binding(GO:0009374) |
| 1.1 | 3.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 1.1 | 3.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 1.1 | 3.2 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 1.0 | 3.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 1.0 | 4.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 1.0 | 3.9 | GO:0050473 | arachidonate 15-lipoxygenase activity(GO:0050473) |
| 1.0 | 6.8 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.9 | 4.7 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.9 | 3.7 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.9 | 3.6 | GO:0016972 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.9 | 2.7 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.9 | 2.7 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.9 | 4.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.8 | 3.2 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.8 | 2.4 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.8 | 4.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.8 | 4.0 | GO:0070905 | serine binding(GO:0070905) |
| 0.8 | 3.2 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.8 | 4.0 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.8 | 2.4 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.8 | 2.4 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.8 | 3.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.8 | 2.3 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.8 | 2.3 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.8 | 8.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.8 | 4.5 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.8 | 3.0 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.8 | 2.3 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.7 | 4.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.7 | 3.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.7 | 2.9 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.7 | 2.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.7 | 1.4 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.7 | 11.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.7 | 2.0 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.7 | 2.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.6 | 3.8 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.6 | 3.2 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.6 | 1.8 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.6 | 4.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.6 | 2.3 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.6 | 2.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.5 | 3.2 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.5 | 2.7 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.5 | 3.7 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.5 | 4.7 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.5 | 5.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.5 | 4.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.5 | 1.5 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.5 | 2.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.5 | 1.4 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.5 | 2.4 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.5 | 1.4 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.5 | 3.8 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.5 | 1.9 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.5 | 3.8 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.5 | 1.9 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.5 | 2.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.5 | 4.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.5 | 1.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.5 | 1.4 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.5 | 8.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.4 | 0.9 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.4 | 1.7 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.4 | 10.1 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.4 | 3.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.4 | 2.5 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.4 | 1.6 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.4 | 2.0 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.4 | 4.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.4 | 1.9 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.4 | 1.6 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.4 | 3.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.4 | 2.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.4 | 4.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.4 | 2.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.4 | 1.9 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.4 | 2.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.4 | 1.1 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.4 | 2.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 8.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.4 | 1.4 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.3 | 1.0 | GO:0050421 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.3 | 1.0 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.3 | 3.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 1.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.3 | 4.8 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.3 | 1.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.3 | 1.0 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.3 | 1.9 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.3 | 2.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.3 | 4.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 8.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.3 | 0.9 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.3 | 6.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 3.1 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.3 | 12.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.3 | 1.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 3.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.3 | 1.2 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.3 | 1.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.3 | 3.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.3 | 2.4 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.3 | 1.2 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.3 | 2.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.3 | 3.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.3 | 2.3 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.3 | 0.8 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.3 | 0.8 | GO:0051379 | beta-adrenergic receptor activity(GO:0004939) epinephrine binding(GO:0051379) norepinephrine binding(GO:0051380) |
| 0.3 | 0.8 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.3 | 0.8 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.3 | 0.8 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.3 | 3.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.3 | 2.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.3 | 1.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.3 | 2.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.3 | 1.8 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.3 | 4.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 0.7 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.2 | 1.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 3.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.2 | 1.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 1.6 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.2 | 1.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.2 | 1.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 17.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.2 | 1.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 1.5 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.2 | 1.5 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 1.5 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.2 | 1.5 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.2 | 0.8 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.2 | 0.2 | GO:0015923 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.2 | 5.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 2.7 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 0.8 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.2 | 4.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 4.9 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.2 | 1.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 0.6 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.2 | 1.7 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 1.7 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.2 | 3.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 1.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.2 | 2.0 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.2 | 0.5 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.2 | 0.9 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 11.4 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.2 | 9.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.2 | 0.7 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.2 | 3.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 1.9 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 4.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 1.0 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.2 | 1.7 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 5.0 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.2 | 7.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 0.6 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 1.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 8.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 1.7 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.2 | 2.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 3.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.6 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 2.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 2.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 2.2 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.1 | 1.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.9 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 0.1 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.1 | 1.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 4.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 4.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 2.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 3.7 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 2.6 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 3.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.7 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 3.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.3 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 3.7 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.5 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.1 | 2.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 3.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.9 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 1.3 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 3.7 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 1.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 2.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 1.7 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 3.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 3.2 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 11.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 18.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 1.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.4 | GO:0032408 | purine-specific mismatch base pair DNA N-glycosylase activity(GO:0000701) MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.1 | 2.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.7 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.1 | 3.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 28.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 4.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.3 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 1.0 | GO:1990380 | Lys63-specific deubiquitinase activity(GO:0061578) Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.4 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.7 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 2.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 9.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 3.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 10.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 1.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 2.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 7.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 4.0 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.3 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.1 | 0.7 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.1 | 3.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 2.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 2.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 1.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 4.7 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.1 | 1.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 2.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.5 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 1.9 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.1 | 12.7 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 0.5 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 1.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.9 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 5.6 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 0.3 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.1 | 3.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 0.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 2.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 1.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.8 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.8 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 1.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.7 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.1 | 0.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.4 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 0.6 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 0.9 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.9 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 1.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.2 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 1.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 2.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 2.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 1.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.4 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 3.6 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.0 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 3.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 8.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 1.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.4 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 1.7 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 7.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 2.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.4 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 4.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.1 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.0 | 20.2 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 1.1 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.4 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 1.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 1.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.0 | GO:0005262 | calcium channel activity(GO:0005262) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 1.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.6 | 2.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.4 | 12.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.4 | 2.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.3 | 1.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.3 | 5.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.3 | 13.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 8.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.2 | 10.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 10.7 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.2 | 8.8 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.2 | 8.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 6.7 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 3.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 2.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 4.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 4.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 35.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 3.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 3.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 5.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.2 | 7.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 4.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.2 | 3.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 25.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 6.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 2.5 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 0.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 4.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 2.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.9 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 6.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 3.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 3.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 4.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 5.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 7.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 4.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 1.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 21.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 4.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 3.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 27.3 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.1 | 3.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 0.7 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 1.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 1.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 0.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 1.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 2.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 3.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.7 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 13.0 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 1.0 | 2.9 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 1.0 | 1.9 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.8 | 1.6 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.8 | 11.8 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.8 | 7.0 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.7 | 0.7 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.6 | 8.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.6 | 1.2 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.5 | 5.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.5 | 13.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.5 | 10.0 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.5 | 12.2 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.4 | 8.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.4 | 5.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.4 | 18.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 4.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 3.8 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.3 | 4.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.3 | 7.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.3 | 4.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.3 | 4.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 2.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.2 | 1.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 2.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 3.5 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.2 | 4.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.2 | 3.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.2 | 13.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 4.5 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 3.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 5.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 2.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.2 | 9.5 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.2 | 2.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 2.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.2 | 3.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 5.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 9.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 2.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 3.0 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 4.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 2.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 2.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 2.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 3.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 5.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 2.5 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.1 | 3.0 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 1.7 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 3.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.0 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 3.9 | REACTOME NEGATIVE REGULATION OF FGFR SIGNALING | Genes involved in Negative regulation of FGFR signaling |
| 0.1 | 6.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 6.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 6.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 4.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 4.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 7.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 2.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 2.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 8.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 10.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 26.1 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.1 | 1.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.8 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 8.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 2.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 2.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 2.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 0.9 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 6.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 0.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 3.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 0.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 0.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 3.2 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 0.7 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 0.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 2.2 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 1.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 2.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 5.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 2.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 2.5 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.8 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.5 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 0.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |