|
chr4_+_126237566
|
8.397
|
NM_024582
|
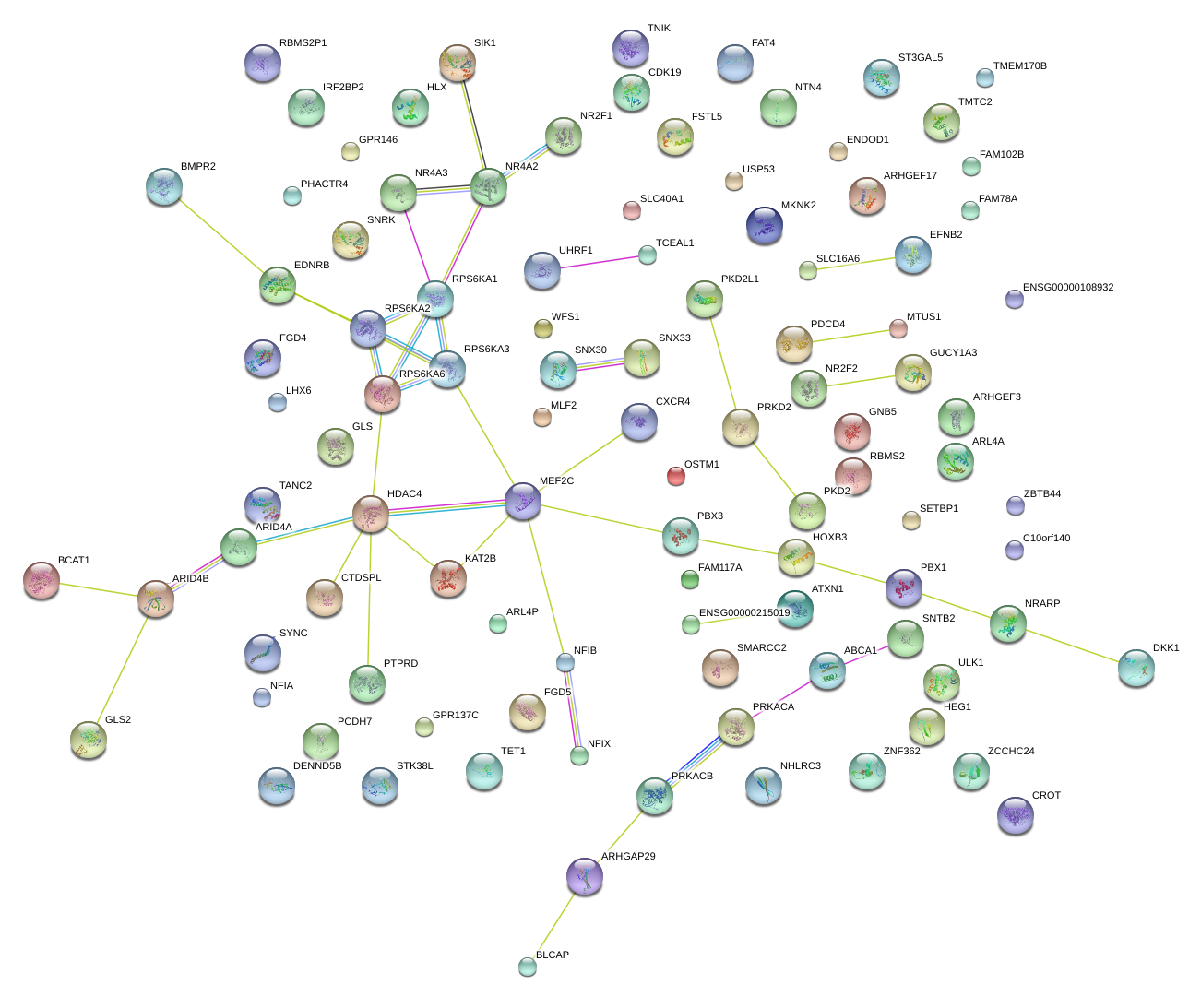
FAT4
|
FAT tumor suppressor homolog 4 (Drosophila)
|
|
chr1_+_61542928
|
7.017
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr2_-_190445521
|
6.991
|
NM_014585
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1
|
|
chr1_+_61547625
|
6.834
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_61547388
|
6.353
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr4_+_156588811
|
6.039
|
NM_001130684
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr12_+_27397077
|
5.688
|
NM_015000
|
STK38L
|
serine/threonine kinase 38 like
|
|
chr10_-_81205091
|
5.665
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr9_-_14398981
|
5.431
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr18_+_42260774
|
5.379
|
NM_015559
|
SETBP1
|
SET binding protein 1
|
|
chr9_-_14314036
|
4.972
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr9_-_134151905
|
4.751
|
NM_033387
|
FAM78A
|
family with sequence similarity 78, member A
|
|
chr3_+_43328001
|
4.625
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr7_+_1097140
|
4.574
|
NM_138445
|
GPR146
|
G protein-coupled receptor 146
|
|
chr3_+_20081523
|
4.401
|
NM_003884
|
KAT2B
|
K(lysine) acetyltransferase 2B
|
|
chr10_+_112631552
|
4.340
|
NM_001199492
NM_014456
NM_145341
|
PDCD4
|
programmed cell death 4 (neoplastic transformation inhibitor)
|
|
chr12_+_32654976
|
4.332
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr9_-_107690526
|
4.310
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr6_+_11538486
|
4.118
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr4_+_6271558
|
3.966
|
NM_001145853
NM_006005
|
WFS1
|
Wolfram syndrome 1 (wolframin)
|
|
chr4_+_156587861
|
3.867
|
NM_000856
NM_001130682
NM_001130686
NM_001130687
NM_001130683
NM_001130685
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr9_-_124984018
|
3.708
|
NM_001242335
|
LHX6
|
LIM homeobox 6
|
|
chr1_-_33168356
|
3.673
|
NM_001161708
NM_030786
|
SYNC
|
syncoilin, intermediate filament protein
|
|
chr8_-_17555158
|
3.634
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr2_-_157189040
|
3.507
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr9_-_10612722
|
3.492
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr15_+_96876568
|
3.444
|
NM_001145157
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr9_-_124990706
|
3.442
|
NM_001242333
|
LHX6
|
LIM homeobox 6
|
|
chr12_-_96184510
|
3.413
|
NM_021229
|
NTN4
|
netrin 4
|
|
chr8_-_17580024
|
3.387
|
NM_001166393
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr9_-_8733893
|
3.362
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr6_-_16761600
|
3.358
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr2_-_240322620
|
3.326
|
NM_006037
|
HDAC4
|
histone deacetylase 4
|
|
chr8_-_17658385
|
3.281
|
NM_001001924
NM_001001925
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr2_+_191745546
|
3.248
|
NM_014905
|
GLS
|
glutaminase
|
|
chr8_-_17579729
|
3.162
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr15_+_96875793
|
3.097
|
NM_001145156
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr9_-_124990949
|
2.984
|
NM_014368
NM_199160
|
LHX6
|
LIM homeobox 6
|
|
chr12_-_31743822
|
2.931
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr1_+_84609941
|
2.929
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr1_+_84630575
|
2.914
|
NM_001242860
NM_001242861
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr1_+_84630014
|
2.885
|
NM_001242857
NM_001242858
NM_001242859
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr15_+_96869156
|
2.858
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr17_+_61086897
|
2.841
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr5_-_88179278
|
2.838
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr9_+_115513050
|
2.835
|
NM_001012994
|
SNX30
|
sorting nexin family member 30
|
|
chr6_-_111136407
|
2.751
|
NM_015076
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr1_+_84647342
|
2.693
|
NM_001242862
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr2_-_86094792
|
2.665
|
NM_001042437
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5
|
|
chr5_-_88199868
|
2.629
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr13_-_78493902
|
2.605
|
NM_001201397
|
EDNRB
|
endothelin receptor type B
|
|
chr4_+_88928797
|
2.563
|
NM_000297
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant)
|
|
chr19_+_4910339
|
2.562
|
NM_013282
|
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chr13_-_78549663
|
2.522
|
NM_000115
|
EDNRB
|
endothelin receptor type B
|
|
chr7_+_86974928
|
2.451
|
NM_001143935
NM_001243745
NM_021151
|
CROT
|
carnitine O-octanoyltransferase
|
|
chr1_-_94703252
|
2.401
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr14_+_53019865
|
2.353
|
NM_001099652
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr15_+_96873845
|
2.329
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr2_-_86116016
|
2.261
|
NM_003896
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5
|
|
chr1_-_234745223
|
2.254
|
NM_001077397
NM_182972
|
IRF2BP2
|
interferon regulatory factor 2 binding protein 2
|
|
chr12_+_132379262
|
2.245
|
NM_003565
|
ULK1
|
unc-51-like kinase 1 (C. elegans)
|
|
chrX_+_102883647
|
2.145
|
NM_001006640
|
TCEAL1
|
transcription elongation factor A (SII)-like 1
|
|
chr1_+_33722158
|
2.112
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr5_-_88119604
|
2.095
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr3_+_37903643
|
2.079
|
NM_001008392
NM_005808
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chr6_-_108395939
|
2.040
|
NM_014028
|
OSTM1
|
osteopetrosis associated transmembrane protein 1
|
|
chr11_-_130184459
|
2.037
|
NM_014155
|
ZBTB44
|
zinc finger and BTB domain containing 44
|
|
chr17_-_47841517
|
2.011
|
NM_030802
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr10_+_54074037
|
1.943
|
NM_012242
|
DKK1
|
dickkopf 1 homolog (Xenopus laevis)
|
|
chr10_+_70320044
|
1.942
|
NM_030625
|
TET1
|
tet methylcytosine dioxygenase 1
|
|
chr12_+_56915587
|
1.937
|
NM_002898
|
RBMS2
|
RNA binding motif, single stranded interacting protein 2
|
|
chr13_-_107187312
|
1.930
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr9_-_124989755
|
1.927
|
NM_001242334
|
LHX6
|
LIM homeobox 6
|
|
chr3_+_14860468
|
1.926
|
NM_152536
|
FGD5
|
FYVE, RhoGEF and PH domain containing 5
|
|
chr19_+_4909447
|
1.909
|
NM_001048201
|
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chrX_+_102883891
|
1.890
|
NM_001006639
NM_004780
|
TCEAL1
|
transcription elongation factor A (SII)-like 1
|
|
chr12_-_25055966
|
1.865
|
NM_001178093
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr12_-_25102281
|
1.848
|
NM_001178091
NM_001178092
NM_005504
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr11_+_94822937
|
1.843
|
NM_015036
|
ENDOD1
|
endonuclease domain containing 1
|
|
chr9_+_128510477
|
1.828
|
NM_001134778
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr13_+_39612440
|
1.812
|
NM_001012754
NM_001017370
|
NHLRC3
|
NHL repeat containing 3
|
|
chr15_-_52483564
|
1.794
|
NM_016194
|
GNB5
|
guanine nucleotide binding protein (G protein), beta 5
|
|
chr12_-_25055228
|
1.786
|
NM_001178094
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr4_+_120133764
|
1.778
|
NM_019050
|
USP53
|
ubiquitin specific peptidase 53
|
|
chr12_+_83080901
|
1.778
|
NM_152588
|
TMTC2
|
transmembrane and tetratricopeptide repeat containing 2
|
|
chr10_-_21814610
|
1.761
|
NM_207371
|
C10orf140
|
chromosome 10 open reading frame 140
|
|
chr1_+_109102970
|
1.748
|
NM_001010883
|
FAM102B
|
family with sequence similarity 102, member B
|
|
chr1_+_221052735
|
1.743
|
NM_021958
|
HLX
|
H2.0-like homeobox
|
|
chr2_+_203241044
|
1.736
|
NM_001204
|
BMPR2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase)
|
|
chr19_-_2051222
|
1.734
|
NM_017572
NM_199054
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chrX_-_20284708
|
1.715
|
NM_004586
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr7_+_12726413
|
1.690
|
NM_001037164
NM_212460
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr3_-_171177851
|
1.675
|
NM_001161560
NM_001161561
NM_001161562
NM_001161563
NM_001161564
NM_001161565
NM_001161566
NM_015028
|
TNIK
|
TRAF2 and NCK interacting kinase
|
|
chr12_-_56583301
|
1.641
|
NM_001130420
NM_003075
NM_139067
|
SMARCC2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2
|
|
chr1_+_28764660
|
1.638
|
NM_023923
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr7_+_12726910
|
1.632
|
NM_001195396
NM_005738
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr9_+_128509616
|
1.631
|
NM_006195
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr2_-_136875629
|
1.619
|
NM_003467
|
CXCR4
|
chemokine (C-X-C motif) receptor 4
|
|
chr20_-_36156082
|
1.574
|
NM_001167822
NM_001167821
|
BLCAP
|
bladder cancer associated protein
|
|
chr3_-_124774735
|
1.543
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr17_-_66287256
|
1.534
|
NM_001174166
NM_004694
|
SLC16A6
|
solute carrier family 16, member 6 (monocarboxylic acid transporter 7)
|
|
chr20_-_36156282
|
1.521
|
NM_001167820
NM_006698
|
BLCAP
|
bladder cancer associated protein
|
|
chr1_+_28696056
|
1.481
|
NM_001048183
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr2_-_136873740
|
1.475
|
NM_001008540
|
CXCR4
|
chemokine (C-X-C motif) receptor 4
|
|
chr21_-_44846927
|
1.474
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr3_-_57113292
|
1.430
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr4_+_30721865
|
1.429
|
NM_032456
NM_001173523
NM_002589
NM_032457
|
PCDH7
|
protocadherin 7
|
|
chr4_-_163085117
|
1.427
|
NM_001128427
NM_001128428
NM_020116
|
FSTL5
|
follistatin-like 5
|
|
chr16_+_69220940
|
1.416
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr20_-_36152952
|
1.387
|
NM_001167823
|
BLCAP
|
bladder cancer associated protein
|
|
chr17_-_46651809
|
1.385
|
NM_002146
|
HOXB3
|
homeobox B3
|
|
chr14_+_58765209
|
1.384
|
NM_002892
NM_023000
NM_023001
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr13_-_78492906
|
1.363
|
NM_001122659
NM_003991
|
EDNRB
|
endothelin receptor type B
|
|
chr11_+_73019662
|
1.307
|
NM_014786
|
ARHGEF17
|
Rho guanine nucleotide exchange factor (GEF) 17
|
|
chr9_-_140196702
|
1.305
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr11_+_9596233
|
1.301
|
NM_001143976
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr4_-_125633716
|
1.301
|
NM_001167882
NM_020337
|
ANKRD50
|
ankyrin repeat domain 50
|
|
chr3_-_56809594
|
1.298
|
NM_001128616
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr19_-_31840118
|
1.263
|
NM_020856
|
TSHZ3
|
teashirt zinc finger homeobox 3
|
|
chr13_+_115047040
|
1.258
|
NM_023011
NM_080687
|
UPF3A
|
UPF3 regulator of nonsense transcripts homolog A (yeast)
|
|
chr2_-_158731622
|
1.257
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr16_+_68119211
|
1.249
|
NM_004555
NM_173163
NM_173165
|
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3
|
|
chr18_-_19284737
|
1.237
|
NM_138340
|
ABHD3
|
abhydrolase domain containing 3
|
|
chr2_+_16080639
|
1.222
|
NM_005378
|
MYCN
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian)
|
|
chr2_-_48115846
|
1.220
|
NM_025133
|
FBXO11
|
F-box protein 11
|
|
chr9_-_16870719
|
1.220
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr3_-_56835829
|
1.207
|
NM_019555
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr2_+_70485204
|
1.197
|
NM_016297
|
PCYOX1
|
prenylcysteine oxidase 1
|
|
chr17_+_38278789
|
1.186
|
NM_001012241
|
MSL1
|
male-specific lethal 1 homolog (Drosophila)
|
|
chr3_+_23987514
|
1.178
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr1_+_33231227
|
1.145
|
NM_001198972
NM_001198973
|
KIAA1522
|
KIAA1522
|
|
chr2_+_178257371
|
1.144
|
NM_003659
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr14_-_75593720
|
1.144
|
NM_033116
|
NEK9
|
NIMA (never in mitosis gene a)- related kinase 9
|
|
chr7_-_105319608
|
1.138
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr16_-_71323090
|
1.128
|
NM_001099642
NM_018348
|
FTSJD1
|
FtsJ methyltransferase domain containing 1
|
|
chr1_+_26872342
|
1.121
|
NM_001006665
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr15_-_52472075
|
1.116
|
NM_006578
|
GNB5
|
guanine nucleotide binding protein (G protein), beta 5
|
|
chr10_+_3110818
|
1.104
|
NM_001242339
|
PFKP
|
phosphofructokinase, platelet
|
|
chr20_-_62601140
|
1.101
|
NM_020713
|
ZNF512B
|
zinc finger protein 512B
|
|
chr5_+_153570270
|
1.092
|
NM_198321
|
GALNT10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10)
|
|
chr9_+_82186846
|
1.085
|
NM_007005
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr8_-_124408688
|
1.083
|
NM_014109
|
ATAD2
|
ATPase family, AAA domain containing 2
|
|
chr1_+_33207343
|
1.062
|
NM_020888
|
KIAA1522
|
KIAA1522
|
|
chr16_+_53088791
|
1.060
|
NM_025134
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr17_+_76000248
|
1.060
|
NM_001142640
NM_018996
|
TNRC6C
|
trinucleotide repeat containing 6C
|
|
chr2_-_158732341
|
1.052
|
NM_001111067
|
ACVR1
|
activin A receptor, type I
|
|
chr2_-_55646946
|
1.052
|
NM_001135597
NM_018084
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr10_-_46090191
|
1.047
|
NM_001002265
NM_145021
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8
|
|
chr17_-_40307015
|
1.034
|
NM_001252039
NM_004583
NM_201434
|
RAB5C
|
RAB5C, member RAS oncogene family
|
|
chr11_-_63536112
|
1.028
|
NM_001144936
|
C11orf95
|
chromosome 11 open reading frame 95
|
|
chr6_-_30585016
|
1.024
|
NM_002714
|
PPP1R10
|
protein phosphatase 1, regulatory subunit 10
|
|
chr1_-_47695442
|
1.021
|
NM_003189
|
TAL1
|
T-cell acute lymphocytic leukemia 1
|
|
chr8_-_57123858
|
1.019
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr11_+_9595183
|
1.019
|
NM_003390
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr18_-_29264394
|
1.012
|
NM_004775
|
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6
|
|
chr6_+_7727010
|
1.005
|
NM_001718
|
BMP6
|
bone morphogenetic protein 6
|
|
chr7_+_100136784
|
1.000
|
NM_006076
|
AGFG2
|
ArfGAP with FG repeats 2
|
|
chr1_+_26856240
|
0.999
|
NM_002953
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr1_-_68299047
|
0.975
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr16_-_47494981
|
0.964
|
NM_030790
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1
|
|
chr6_+_119215240
|
0.957
|
NM_014034
|
ASF1A
|
ASF1 anti-silencing function 1 homolog A (S. cerevisiae)
|
|
chr2_-_197036301
|
0.946
|
NM_004226
|
STK17B
|
serine/threonine kinase 17b
|
|
chr13_+_78271820
|
0.945
|
NM_001242868
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr2_+_204192915
|
0.943
|
NM_005759
|
ABI2
|
abl-interactor 2
|
|
chr11_-_102323353
|
0.939
|
NM_052932
|
TMEM123
|
transmembrane protein 123
|
|
chr7_+_106809405
|
0.933
|
NM_001244262
NM_012257
|
HBP1
|
HMG-box transcription factor 1
|
|
chr14_-_35183722
|
0.932
|
NM_001243645
NM_138638
|
CFL2
|
cofilin 2 (muscle)
|
|
chr2_-_69870976
|
0.926
|
NM_014911
|
AAK1
|
AP2 associated kinase 1
|
|
chr10_-_91295310
|
0.907
|
NM_213606
|
SLC16A12
|
solute carrier family 16, member 12 (monocarboxylic acid transporter 12)
|
|
chr9_+_36036900
|
0.900
|
NM_021111
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs
|
|
chr1_-_156051788
|
0.899
|
NM_001093725
|
MEX3A
|
mex-3 homolog A (C. elegans)
|
|
chr3_+_156392714
|
0.897
|
NM_001184717
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr3_-_145968958
|
0.888
|
NM_001128306
NM_001177304
NM_020353
|
PLSCR4
|
phospholipid scramblase 4
|
|
chr4_-_76439582
|
0.887
|
NM_001008925
NM_001009922
NM_015436
|
RCHY1
|
ring finger and CHY zinc finger domain containing 1
|
|
chr8_-_91657898
|
0.884
|
NM_001008495
NM_001146273
|
TMEM64
|
transmembrane protein 64
|
|
chr10_-_46030798
|
0.883
|
NM_001002266
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8
|
|
chr9_-_127952037
|
0.879
|
NM_001123355
NM_001123369
NM_002721
|
PPP6C
|
protein phosphatase 6, catalytic subunit
|
|
chr3_-_197463713
|
0.879
|
NM_014687
|
KIAA0226
|
KIAA0226
|
|
chr11_+_61520000
|
0.877
|
NM_001127392
|
C11orf9
|
chromosome 11 open reading frame 9
|
|
chr13_+_78273015
|
0.866
|
NM_001040153
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr10_+_102295572
|
0.857
|
NM_017902
|
HIF1AN
|
hypoxia inducible factor 1, alpha subunit inhibitor
|
|
chr4_+_123844210
|
0.854
|
NM_145207
|
SPATA5
|
spermatogenesis associated 5
|
|
chr12_-_118406027
|
0.851
|
NM_173598
|
KSR2
|
kinase suppressor of ras 2
|
|
chr7_+_86781869
|
0.840
|
NM_021145
|
DMTF1
|
cyclin D binding myb-like transcription factor 1
|
|
chr3_+_23986735
|
0.820
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr17_-_35969401
|
0.818
|
NM_001163544
NM_001163545
NM_001163546
NM_001163547
NM_007247
NM_080550
NM_198882
|
SYNRG
|
synergin, gamma
|
|
chr19_+_14016955
|
0.809
|
NM_017721
|
CC2D1A
|
coiled-coil and C2 domain containing 1A
|
|
chr10_+_65281108
|
0.807
|
NM_001001330
|
REEP3
|
receptor accessory protein 3
|
|
chr14_-_35182944
|
0.796
|
NM_021914
|
CFL2
|
cofilin 2 (muscle)
|
|
chr8_-_95961543
|
0.795
|
NM_001135733
NM_033285
|
TP53INP1
|
tumor protein p53 inducible nuclear protein 1
|
|
chr2_-_218808705
|
0.795
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr1_-_85155885
|
0.794
|
NM_001166295
NM_001166417
NM_001166293
NM_001166294
NM_014021
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein
|
|
chr2_+_172378856
|
0.787
|
NM_001127383
NM_024843
|
CYBRD1
|
cytochrome b reductase 1
|
|
chr6_+_110501591
|
0.785
|
NM_015891
|
CDC40
|
cell division cycle 40 homolog (S. cerevisiae)
|
|
chr10_+_23728197
|
0.785
|
NM_001145373
|
OTUD1
|
OTU domain containing 1
|
|
chr3_-_13114547
|
0.781
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr10_+_97803064
|
0.774
|
NM_001134375
NM_001134376
NM_019084
|
CCNJ
|
cyclin J
|
|
chr3_-_197476567
|
0.769
|
NM_001145642
|
KIAA0226
|
KIAA0226
|
|
chr3_+_156392204
|
0.752
|
NM_015508
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr20_+_1874779
|
0.752
|
NM_001040022
|
SIRPA
|
signal-regulatory protein alpha
|