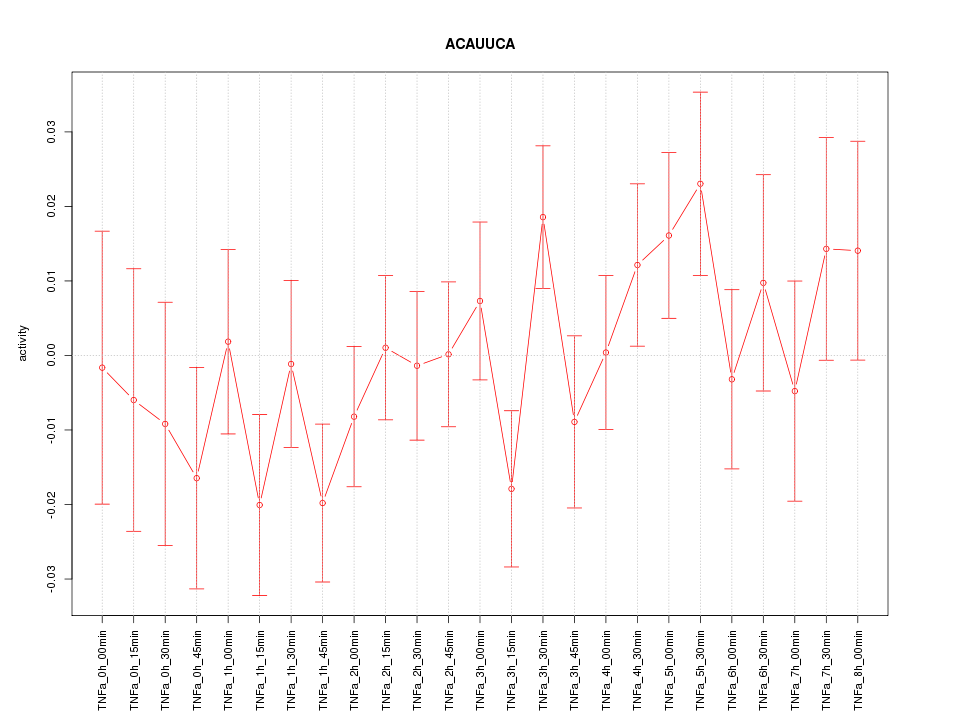
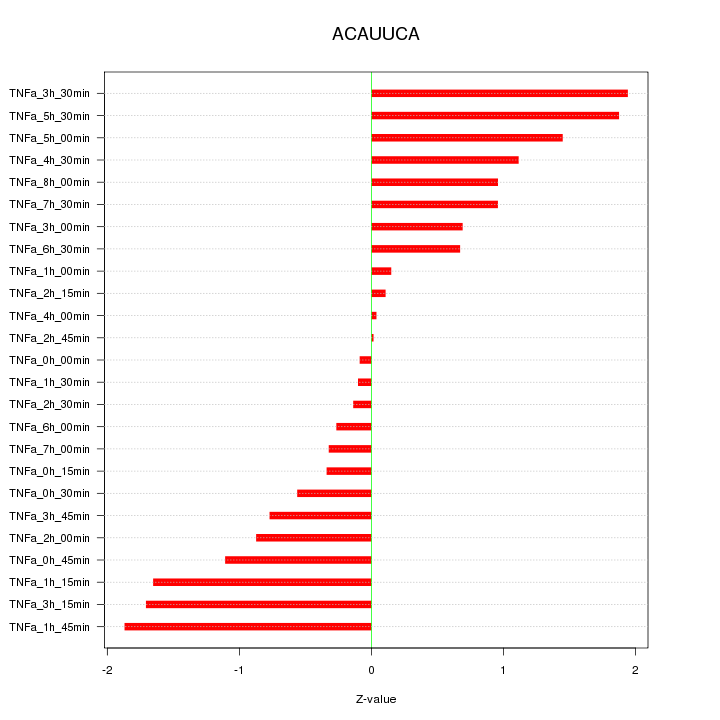
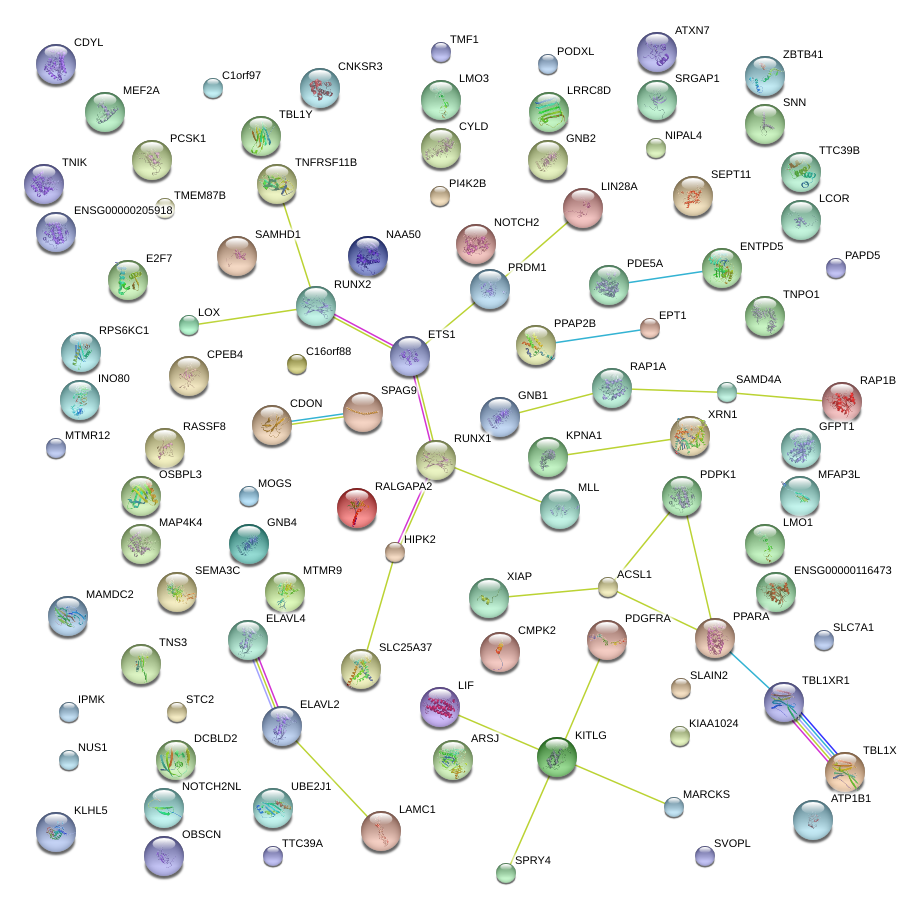
|
chr4_-_120549115
|
2.765
|
NM_033437
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr7_-_25019625
|
2.589
|
NM_015550
NM_145320
NM_145321
NM_145322
|
OSBPL3
|
oxysterol binding protein-like 3
|
|
chr7_-_47621741
|
2.555
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr4_-_120549827
|
2.118
|
NM_001083
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr8_+_23386362
|
1.806
|
NM_016612
|
SLC25A37
|
solute carrier family 25, member 37
|
|
chr11_-_125933186
|
1.757
|
NM_001243597
NM_016952
|
CDON
|
Cdon homolog (mouse)
|
|
chr9_+_72658496
|
1.730
|
NM_153267
|
MAMDC2
|
MAM domain containing 2
|
|
chr4_-_120548329
|
1.699
|
NM_033430
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr5_-_141704575
|
1.620
|
NM_001127496
NM_030964
|
SPRY4
|
sprouty homolog 4 (Drosophila)
|
|
chr1_+_169075869
|
1.402
|
NM_001677
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr5_-_121414011
|
1.337
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr5_-_121412805
|
1.315
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr20_-_20693122
|
1.264
|
NM_020343
|
RALGAPA2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic)
|
|
chr4_-_114900802
|
1.242
|
NM_024590
|
ARSJ
|
arylsulfatase family, member J
|
|
chr2_-_69614321
|
1.234
|
NM_001244710
NM_002056
|
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1
|
|
chr21_-_36421586
|
1.155
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr7_-_131241321
|
1.101
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr8_-_119964059
|
1.097
|
NM_002546
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b
|
|
chr1_+_90286469
|
1.090
|
NM_001134479
|
LRRC8D
|
leucine rich repeat containing 8 family, member D
|
|
chr11_+_118307072
|
1.076
|
NM_001197104
NM_005933
|
MLL
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)
|
|
chr16_+_50776012
|
1.054
|
NM_001042412
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr16_+_50775960
|
1.051
|
NM_001042355
NM_015247
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr12_+_64238511
|
1.043
|
NM_020762
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1
|
|
chrX_+_9432980
|
1.005
|
NM_005647
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr1_+_90287404
|
0.961
|
NM_018103
|
LRRC8D
|
leucine rich repeat containing 8 family, member D
|
|
chr15_+_100106132
|
0.957
|
NM_001130926
NM_001130927
NM_001171894
NM_005587
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr6_-_154831704
|
0.935
|
NM_173515
|
CNKSR3
|
CNKSR family member 3
|
|
chr11_-_128457452
|
0.893
|
NM_001143820
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr5_+_173315313
|
0.892
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr3_-_192635933
|
0.864
|
NM_178496
|
MB21D2
|
Mab-21 domain containing 2
|
|
chr15_+_100173182
|
0.836
|
NM_001130928
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr4_+_55095263
|
0.800
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chrX_+_9431314
|
0.798
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr22_+_46546498
|
0.796
|
NM_001001928
NM_005036
|
PPARA
|
peroxisome proliferator-activated receptor alpha
|
|
chr11_-_128392061
|
0.794
|
NM_001162422
NM_005238
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr15_+_79724857
|
0.787
|
NM_015206
|
KIAA1024
|
KIAA1024
|
|
chr2_-_7005764
|
0.768
|
NM_207315
|
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial
|
|
chr10_+_98592658
|
0.748
|
NM_032440
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr7_-_139477437
|
0.746
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr3_-_98620452
|
0.745
|
NM_080927
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr1_-_120612301
|
0.743
|
NM_001200001
NM_024408
|
NOTCH2
|
notch 2
|
|
chr3_-_142166782
|
0.742
|
NM_001042604
NM_019001
|
XRN1
|
5'-3' exoribonuclease 1
|
|
chr14_+_55221505
|
0.735
|
NM_001161577
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr12_+_26126687
|
0.734
|
NM_001164747
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr20_-_35580186
|
0.732
|
NM_015474
|
SAMHD1
|
SAM domain and HD domain 1
|
|
chr14_+_55034329
|
0.731
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr17_-_49198084
|
0.730
|
NM_001130527
NM_001130528
NM_003971
|
SPAG9
|
sperm associated antigen 9
|
|
chr3_-_69101236
|
0.730
|
NM_007114
|
TMF1
|
TATA element modulatory factor 1
|
|
chr12_+_26205490
|
0.730
|
NM_001164746
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr4_+_48343502
|
0.722
|
NM_020846
|
SLAIN2
|
SLAIN motif family, member 2
|
|
chr4_-_185747183
|
0.698
|
NM_001995
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr4_+_77870864
|
0.694
|
NM_018243
|
SEPT11
|
septin 11
|
|
chr10_+_98592015
|
0.692
|
NM_001170765
NM_001170766
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr12_-_88974094
|
0.688
|
NM_000899
NM_003994
|
KITLG
|
KIT ligand
|
|
chr4_+_25235651
|
0.656
|
NM_018323
|
PI4K2B
|
phosphatidylinositol 4-kinase type 2 beta
|
|
chr12_+_69004614
|
0.652
|
NM_001010942
NM_001251917
NM_001251918
NM_001251921
NM_001251922
NM_015646
|
RAP1B
|
RAP1B, member of RAS oncogene family
|
|
chr2_+_64681270
|
0.650
|
NM_014181
|
LGALSL
|
lectin, galactoside-binding-like
|
|
chr9_-_15250113
|
0.641
|
NM_001168342
|
TTC39B
|
tetratricopeptide repeat domain 39B
|
|
chr3_-_113465101
|
0.629
|
NM_025146
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr3_+_63850181
|
0.628
|
NM_000333
|
ATXN7
|
ataxin 7
|
|
chr9_-_15307325
|
0.625
|
NM_001168339
NM_001168340
NM_001168341
NM_152574
|
TTC39B
|
tetratricopeptide repeat domain 39B
|
|
chr5_-_32313069
|
0.622
|
NM_001040446
|
MTMR12
|
myotubularin related protein 12
|
|
chr2_+_112812799
|
0.619
|
NM_032824
|
TMEM87B
|
transmembrane protein 87B
|
|
chr22_-_30642683
|
0.619
|
NM_002309
|
LIF
|
leukemia inhibitory factor (cholinergic differentiation factor)
|
|
chr1_+_26737232
|
0.602
|
NM_024674
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chrX_+_122993536
|
0.601
|
NM_001204401
NM_001167
|
XIAP
|
X-linked inhibitor of apoptosis
|
|
chr7_-_80548666
|
0.596
|
NM_006379
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr5_-_172756505
|
0.585
|
NM_003714
|
STC2
|
stanniocalcin 2
|
|
chr2_+_26568900
|
0.580
|
NM_033505
|
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific)
|
|
chr5_-_95748688
|
0.580
|
NM_001177876
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr16_+_2587951
|
0.574
|
NM_002613
NM_031268
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr4_-_170924911
|
0.571
|
NM_001009554
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr4_+_39046198
|
0.565
|
NM_001007075
NM_001171654
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr16_+_11762235
|
0.564
|
NM_003498
|
SNN
|
stannin
|
|
chr6_+_114178516
|
0.558
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr2_+_102314162
|
0.556
|
NM_001242560
NM_004834
NM_145686
NM_001242559
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr6_+_106534188
|
0.554
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr12_-_16759531
|
0.548
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr1_+_213224530
|
0.541
|
NM_001136138
NM_012424
|
RPS6KC1
|
ribosomal protein S6 kinase, 52kDa, polypeptide 1
|
|
chr3_+_63884074
|
0.541
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr1_+_50571963
|
0.538
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr5_-_95768660
|
0.537
|
NM_000439
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr6_+_4890225
|
0.533
|
NM_001143971
|
CDYL
|
chromodomain protein, Y-like
|
|
chr1_+_50574589
|
0.531
|
NM_001144774
NM_021952
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr1_+_182992329
|
0.531
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr6_+_4776641
|
0.530
|
NM_004824
|
CDYL
|
chromodomain protein, Y-like
|
|
chr16_+_50186809
|
0.528
|
NM_001040284
NM_001040285
|
PAPD5
|
PAP associated domain containing 5
|
|
chr5_-_95767862
|
0.528
|
NM_001177875
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr3_-_122233781
|
0.527
|
NM_002264
|
KPNA1
|
karyopherin alpha 1 (importin alpha 5)
|
|
chr6_+_4836331
|
0.515
|
NM_001143970
|
CDYL
|
chromodomain protein, Y-like
|
|
chr2_-_74692534
|
0.513
|
NM_001146158
NM_006302
|
MOGS
|
mannosyl-oligosaccharide glucosidase
|
|
chr5_+_72112417
|
0.511
|
NM_002270
|
TNPO1
|
transportin 1
|
|
chr15_-_41408329
|
0.509
|
NM_017553
|
INO80
|
INO80 homolog (S. cerevisiae)
|
|
chr12_-_77459198
|
0.507
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr6_+_117996616
|
0.506
|
NM_138459
|
NUS1
|
nuclear undecaprenyl pyrophosphate synthase 1 homolog (S. cerevisiae)
|
|
chr5_+_156887026
|
0.505
|
NM_001099287
NM_001172292
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chr13_-_30169795
|
0.504
|
NM_003045
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1
|
|
chr1_+_171454636
|
0.503
|
NM_015172
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chr1_-_51787881
|
0.502
|
NM_001080494
|
TTC39A
|
tetratricopeptide repeat domain 39A
|
|
chr10_-_60027630
|
0.500
|
NM_152230
|
IPMK
|
inositol polyphosphate multikinase
|
|
chr8_+_11141983
|
0.498
|
NM_015458
|
MTMR9
|
myotubularin related protein 9
|
|
chr3_+_63953419
|
0.491
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr1_-_197169635
|
0.491
|
NM_194314
|
ZBTB41
|
zinc finger and BTB domain containing 41
|
|
chr3_-_179169330
|
0.488
|
NM_021629
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr1_-_57045235
|
0.483
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr6_-_90062610
|
0.481
|
NM_016021
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr1_-_51810693
|
0.480
|
NM_001144832
|
TTC39A
|
tetratricopeptide repeat domain 39A
|
|
chr12_-_101801499
|
0.478
|
NM_001177
|
ARL1
|
ADP-ribosylation factor-like 1
|
|
chr1_+_167298280
|
0.472
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr4_+_39063851
|
0.472
|
NM_015990
NM_199039
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr3_-_71632741
|
0.468
|
NM_001244808
NM_001012505
NM_001244810
NM_032682
|
FOXP1
|
forkhead box P1
|
|
chr2_+_23608297
|
0.458
|
NM_052920
|
KLHL29
|
kelch-like 29 (Drosophila)
|
|
chr12_-_16760935
|
0.458
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr3_+_53195214
|
0.453
|
NM_006254
NM_212539
|
PRKCD
|
protein kinase C, delta
|
|
chr1_-_205912546
|
0.448
|
NM_052934
NM_134325
|
SLC26A9
|
solute carrier family 26, member 9
|
|
chr12_+_62654120
|
0.448
|
NM_001252078
NM_001252079
NM_006313
|
USP15
|
ubiquitin specific peptidase 15
|
|
chr16_-_3285359
|
0.447
|
NM_001145448
NM_198088
NM_001145446
NM_198087
|
ZNF200
|
zinc finger protein 200
|
|
chr2_-_145277879
|
0.444
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr3_+_42132707
|
0.443
|
NM_001042646
|
TRAK1
|
trafficking protein, kinesin binding 1
|
|
chr7_+_6144504
|
0.442
|
NM_032172
|
USP42
|
ubiquitin specific peptidase 42
|
|
chr5_+_65017993
|
0.438
|
NM_020726
|
NLN
|
neurolysin (metallopeptidase M3 family)
|
|
chr2_+_111878450
|
0.437
|
NM_001204106
NM_001204107
NM_001204108
NM_001204109
NM_001204110
NM_001204111
NM_001204112
NM_001204113
NM_006538
NM_138621
NM_138622
NM_138623
NM_138624
NM_138625
NM_138626
NM_138627
NM_207002
NM_207003
|
BCL2L11
|
BCL2-like 11 (apoptosis facilitator)
|
|
chr16_-_1429612
|
0.435
|
NM_001193389
NM_023076
|
UNKL
|
unkempt homolog (Drosophila)-like
|
|
chr14_+_74111577
|
0.434
|
NM_001201366
NM_031427
|
DNAL1
|
dynein, axonemal, light chain 1
|
|
chr19_-_16683188
|
0.433
|
NM_024881
|
SLC35E1
|
solute carrier family 35, member E1
|
|
chr11_-_16424391
|
0.433
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr15_+_45879416
|
0.431
|
NM_012388
|
PLDN
|
pallidin homolog (mouse)
|
|
chr5_+_72143929
|
0.428
|
NM_153188
|
TNPO1
|
transportin 1
|
|
chr3_-_160283361
|
0.427
|
NM_002268
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3)
|
|
chr21_-_36260972
|
0.424
|
NM_001001890
NM_001122607
|
RUNX1
|
runt-related transcription factor 1
|
|
chr8_-_74791098
|
0.423
|
NM_001001481
NM_018299
|
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative)
|
|
chr2_-_224702300
|
0.422
|
NM_001039569
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit
|
|
chr11_-_85780105
|
0.419
|
NM_001008660
NM_001206946
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr1_+_50569581
|
0.418
|
NM_001144776
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr14_+_70078309
|
0.418
|
NM_014734
|
KIAA0247
|
KIAA0247
|
|
chr15_+_66679107
|
0.413
|
NM_002755
|
MAP2K1
|
mitogen-activated protein kinase kinase 1
|
|
chrX_+_136648313
|
0.411
|
NM_003413
|
ZIC3
|
Zic family member 3
|
|
chr17_+_27717942
|
0.410
|
NM_020791
NM_025142
|
TAOK1
|
TAO kinase 1
|
|
chr12_-_24715349
|
0.408
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr10_-_120514669
|
0.408
|
NM_153810
|
C10orf46
|
chromosome 10 open reading frame 46
|
|
chr5_+_71403040
|
0.402
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr12_-_48499415
|
0.400
|
NM_014554
|
SENP1
|
SUMO1/sentrin specific peptidase 1
|
|
chr17_+_26662547
|
0.399
|
NM_021137
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial)
|
|
chr18_+_55102777
|
0.389
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr3_+_171757413
|
0.389
|
NM_022763
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr14_+_32798478
|
0.388
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr12_-_57030083
|
0.388
|
NM_013449
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A
|
|
chr15_-_65670325
|
0.384
|
NM_004884
|
IGDCC3
|
immunoglobulin superfamily, DCC subclass, member 3
|
|
chr20_+_37101457
|
0.384
|
NM_020336
|
RALGAPB
|
Ral GTPase activating protein, beta subunit (non-catalytic)
|
|
chr12_-_89918531
|
0.383
|
NM_003774
|
GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4)
|
|
chr10_+_102732285
|
0.381
|
NM_001203244
NM_017893
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr6_+_138483024
|
0.380
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr1_+_50513685
|
0.375
|
NM_001144777
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr4_-_41750936
|
0.370
|
NM_003924
|
PHOX2B
|
paired-like homeobox 2b
|
|
chr12_+_108523464
|
0.369
|
NM_014653
|
WSCD2
|
WSC domain containing 2
|
|
chr15_-_40401041
|
0.368
|
NM_001003940
|
BMF
|
Bcl2 modifying factor
|
|
chr12_-_56652118
|
0.367
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr6_+_106546736
|
0.363
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr2_-_225450105
|
0.362
|
NM_003590
|
CUL3
|
cullin 3
|
|
chr5_+_34929679
|
0.361
|
NM_001012339
NM_194283
|
DNAJC21
|
DnaJ (Hsp40) homolog, subfamily C, member 21
|
|
chr2_+_36583369
|
0.358
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr3_+_88188261
|
0.356
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr4_+_7045142
|
0.355
|
NM_152293
|
TADA2B
|
transcriptional adaptor 2B
|
|
chr15_+_63481713
|
0.354
|
NM_016530
|
RAB8B
|
RAB8B, member RAS oncogene family
|
|
chr13_+_96329392
|
0.351
|
NM_006260
|
DNAJC3
|
DnaJ (Hsp40) homolog, subfamily C, member 3
|
|
chr10_+_134351272
|
0.348
|
NM_005539
|
INPP5A
|
inositol polyphosphate-5-phosphatase, 40kDa
|
|
chr2_-_197457250
|
0.346
|
NM_020760
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2
|
|
chr19_+_40697516
|
0.344
|
NM_002446
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10
|
|
chr7_+_97736118
|
0.344
|
NM_014916
|
LMTK2
|
lemur tyrosine kinase 2
|
|
chr3_-_11623829
|
0.340
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr5_-_142783975
|
0.339
|
NM_001018076
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr4_-_76555570
|
0.339
|
NM_003948
|
CDKL2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase)
|
|
chr16_+_20911556
|
0.338
|
NM_020424
|
LYRM1
|
LYR motif containing 1
|
|
chr3_-_50540853
|
0.338
|
NM_001005505
NM_001174051
NM_006030
|
CACNA2D2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2
|
|
chr2_-_206950895
|
0.337
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr7_+_103969103
|
0.337
|
NM_199000
|
LHFPL3
|
lipoma HMGIC fusion partner-like 3
|
|
chr11_+_32914359
|
0.334
|
NM_001076786
|
QSER1
|
glutamine and serine rich 1
|
|
chr6_+_34759743
|
0.334
|
NM_017754
|
UHRF1BP1
|
UHRF1 binding protein 1
|
|
chr16_+_20911987
|
0.333
|
NM_001128301
NM_001128302
|
LYRM1
|
LYR motif containing 1
|
|
chr4_-_170947428
|
0.332
|
NM_021647
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr11_+_10326618
|
0.331
|
NM_001124
|
ADM
|
adrenomedullin
|
|
chr8_-_124054591
|
0.329
|
NM_001134671
NM_024295
|
DERL1
|
Der1-like domain family, member 1
|
|
chr2_-_225907312
|
0.329
|
NM_014689
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr12_+_12764755
|
0.327
|
NM_001310
|
CREBL2
|
cAMP responsive element binding protein-like 2
|
|
chr11_-_85780899
|
0.325
|
NM_001206947
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr2_-_201936389
|
0.324
|
NM_173822
|
FAM126B
|
family with sequence similarity 126, member B
|
|
chr16_-_3285069
|
0.323
|
NM_001145447
NM_003454
|
ZNF200
|
zinc finger protein 200
|
|
chr9_+_101867401
|
0.322
|
NM_001130916
NM_004612
|
TGFBR1
|
transforming growth factor, beta receptor 1
|
|
chr2_-_113542970
|
0.320
|
NM_000575
|
IL1A
|
interleukin 1, alpha
|
|
chr12_-_498428
|
0.320
|
NM_001042603
|
KDM5A
|
lysine (K)-specific demethylase 5A
|
|
chr6_+_43211221
|
0.318
|
NM_032538
|
TTBK1
|
tau tubulin kinase 1
|
|
chr15_-_40398573
|
0.316
|
NM_033503
|
BMF
|
Bcl2 modifying factor
|
|
chr12_+_26111951
|
0.313
|
NM_001164748
NM_007211
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr14_-_61190458
|
0.313
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr20_+_32150139
|
0.313
|
NM_001039709
NM_005093
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr8_+_61591239
|
0.311
|
NM_017780
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr17_-_4167028
|
0.311
|
NM_016376
NM_020740
|
ANKFY1
|
ankyrin repeat and FYVE domain containing 1
|
|
chr7_+_18126543
|
0.308
|
NM_001204144
|
HDAC9
|
histone deacetylase 9
|
|
chr3_-_11610264
|
0.308
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr5_-_158634641
|
0.305
|
NM_001199382
|
RNF145
|
ring finger protein 145
|