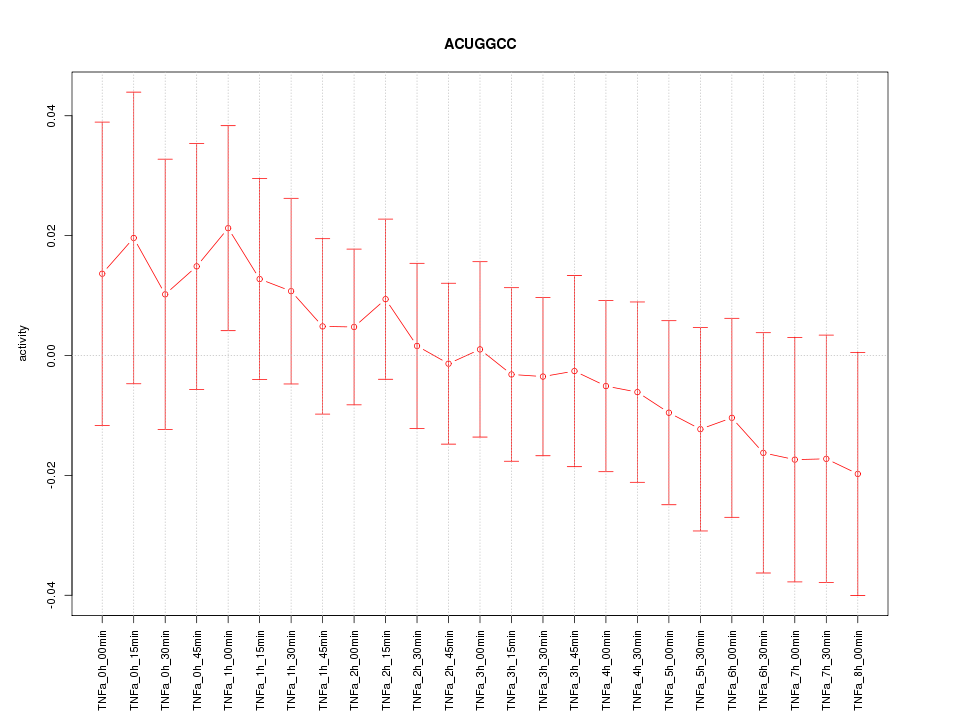
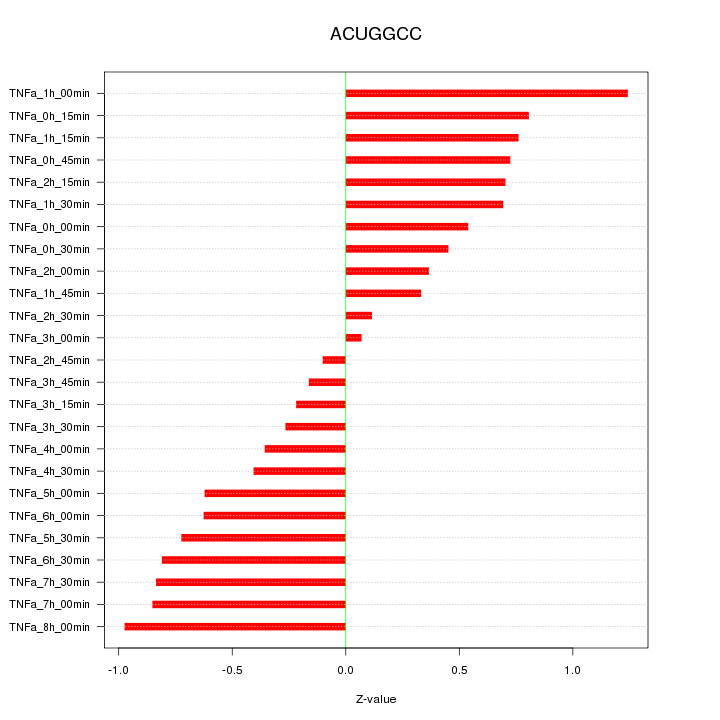
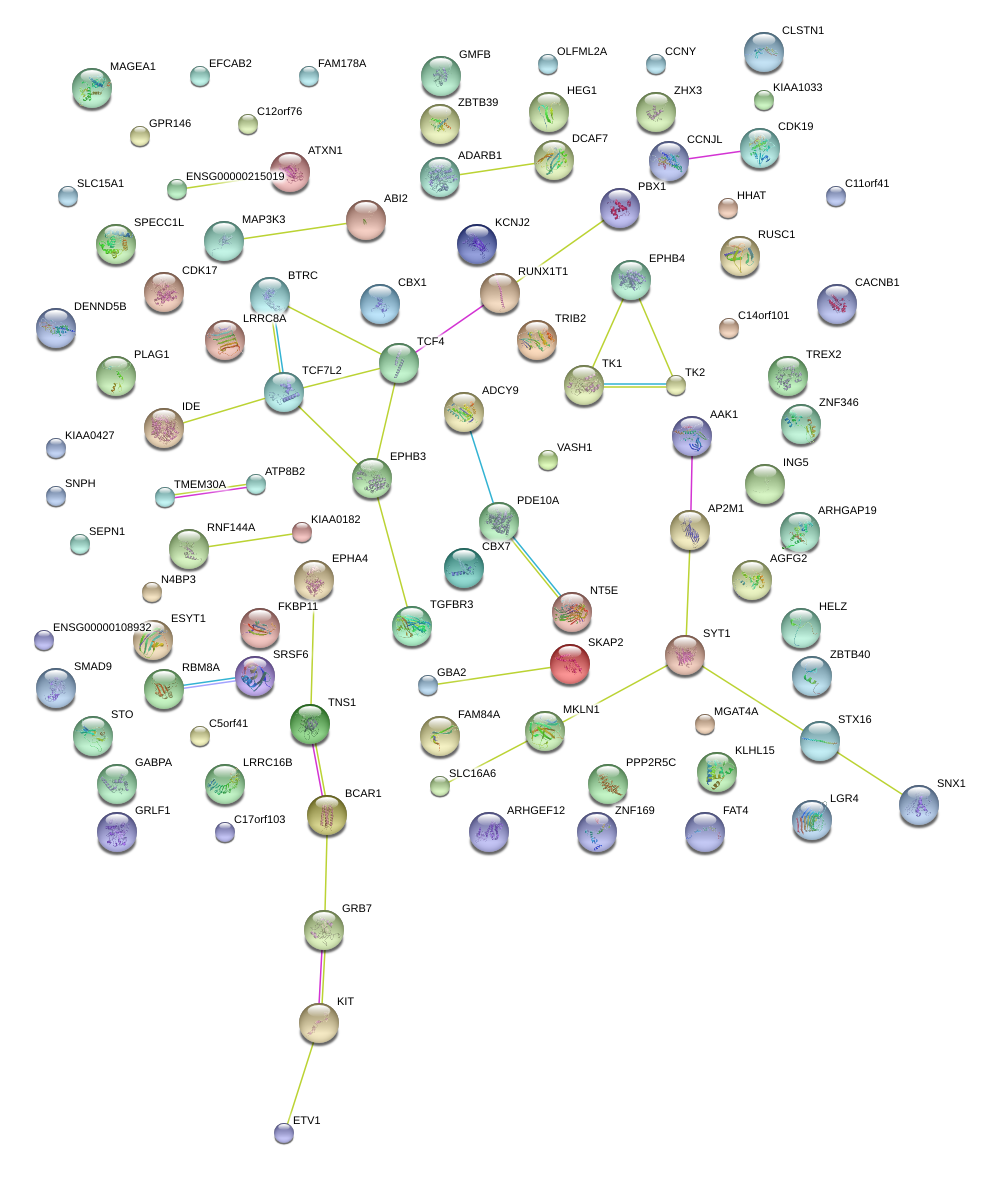
|
chr1_-_92371558
|
2.138
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr8_-_93029864
|
2.021
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_-_92351613
|
2.014
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr8_-_93107881
|
1.894
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr4_+_55523906
|
1.869
|
NM_000222
NM_001093772
|
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
|
|
chr8_-_93111915
|
1.865
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93107442
|
1.797
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr17_+_68165660
|
1.716
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr2_+_12856813
|
1.696
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr8_-_93115453
|
1.634
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr7_+_1097140
|
1.540
|
NM_138445
|
GPR146
|
G protein-coupled receptor 146
|
|
chr8_-_93075190
|
1.502
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr9_-_35749209
|
1.287
|
NM_020944
|
GBA2
|
glucosidase, beta (bile acid) 2
|
|
chr4_+_126237566
|
1.197
|
NM_024582
|
FAT4
|
FAT tumor suppressor homolog 4 (Drosophila)
|
|
chr12_+_79257772
|
1.134
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr12_+_79439432
|
1.124
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr12_+_79258448
|
1.065
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr22_-_39548448
|
0.956
|
NM_175709
|
CBX7
|
chromobox homolog 7
|
|
chr6_+_86159301
|
0.928
|
NM_001204813
NM_002526
|
NT5E
|
5'-nucleotidase, ecto (CD73)
|
|
chr2_+_7057471
|
0.764
|
NM_014746
|
RNF144A
|
ring finger protein 144A
|
|
chr12_-_31743822
|
0.754
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr2_+_204192915
|
0.692
|
NM_005759
|
ABI2
|
abl-interactor 2
|
|
chr3_-_124774735
|
0.679
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr1_+_164528586
|
0.656
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr11_+_33563772
|
0.597
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chr16_-_4166185
|
0.583
|
NM_001116
|
ADCY9
|
adenylate cyclase 9
|
|
chr14_-_54955714
|
0.579
|
NM_004124
|
GMFB
|
glia maturation factor, beta
|
|
chr2_-_222436996
|
0.513
|
NM_004438
|
EPHA4
|
EPH receptor A4
|
|
chr17_+_61699786
|
0.493
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr9_+_131644780
|
0.420
|
NM_001127244
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr17_-_66287256
|
0.407
|
NM_001174166
NM_004694
|
SLC16A6
|
solute carrier family 16, member 6 (monocarboxylic acid transporter 7)
|
|
chr18_-_53255734
|
0.402
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr7_-_26904047
|
0.397
|
NM_003930
|
SKAP2
|
src kinase associated phosphoprotein 2
|
|
chr11_-_27494232
|
0.392
|
NM_018490
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4
|
|
chr5_+_176449696
|
0.390
|
NM_012279
|
ZNF346
|
zinc finger protein 346
|
|
chr9_+_131644368
|
0.390
|
NM_001127245
NM_019594
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr7_-_14031049
|
0.388
|
NM_001163148
|
ETV1
|
ets variant 1
|
|
chr7_-_14029641
|
0.376
|
NM_001163147
NM_004956
|
ETV1
|
ets variant 1
|
|
chr17_+_37895023
|
0.374
|
NM_001242442
|
GRB7
|
growth factor receptor-bound protein 7
|
|
chr6_-_111136407
|
0.368
|
NM_015076
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr6_-_16761600
|
0.357
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr21_+_27107322
|
0.331
|
NM_002040
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa
|
|
chr20_+_1246959
|
0.330
|
NM_014723
|
SNPH
|
syntaphilin
|
|
chr14_+_77228132
|
0.330
|
NM_014909
|
VASH1
|
vasohibin 1
|
|
chr17_+_37894558
|
0.329
|
NM_001030002
|
GRB7
|
growth factor receptor-bound protein 7
|
|
chr21_+_27107257
|
0.329
|
NM_001197297
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa
|
|
chr14_+_57046490
|
0.329
|
NM_017799
|
C14orf101
|
chromosome 14 open reading frame 101
|
|
chr6_-_75994631
|
0.312
|
NM_001143958
NM_018247
|
TMEM30A
|
transmembrane protein 30A
|
|
chr11_+_120207617
|
0.306
|
NM_001198665
NM_015313
|
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12
|
|
chr10_-_99052429
|
0.298
|
NM_001204300
NM_032900
|
ARHGAP19
|
Rho GTPase activating protein 19
|
|
chr18_+_46065367
|
0.297
|
NM_001142397
NM_014772
|
CTIF
|
CBP80/20-dependent translation initiation factor
|
|
chr13_-_99404883
|
0.296
|
NM_005073
|
SLC15A1
|
solute carrier family 15 (oligopeptide transporter), member 1
|
|
chr2_+_242641310
|
0.278
|
NM_032329
|
ING5
|
inhibitor of growth family, member 5
|
|
chr8_-_57123858
|
0.256
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr7_-_14025826
|
0.254
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr10_-_94333847
|
0.249
|
NM_004969
|
IDE
|
insulin-degrading enzyme
|
|
chr17_+_37894161
|
0.249
|
NM_005310
|
GRB7
|
growth factor receptor-bound protein 7
|
|
chr1_+_210502188
|
0.248
|
NM_001170587
NM_001170588
NM_018194
|
HHAT
|
hedgehog acyltransferase
|
|
chr17_+_37896219
|
0.240
|
NM_001242443
|
GRB7
|
growth factor receptor-bound protein 7
|
|
chr5_-_159739482
|
0.236
|
NM_024565
|
CCNJL
|
cyclin J-like
|
|
chr2_-_99347588
|
0.234
|
NM_012214
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr3_+_183892569
|
0.229
|
NM_001025205
NM_004068
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr7_-_14030864
|
0.228
|
NM_001163149
|
ETV1
|
ets variant 1
|
|
chr1_-_9884549
|
0.224
|
NM_001009566
NM_014944
|
CLSTN1
|
calsyntenin 1
|
|
chr20_+_57226287
|
0.223
|
NM_001001433
NM_001134772
NM_001134773
NM_001204868
NM_003763
|
STX16-NPEPL1
STX16
|
STX16-NPEPL1 readthrough (non-protein coding)
syntaxin 16
|
|
chr9_+_127539382
|
0.218
|
NM_182487
|
OLFML2A
|
olfactomedin-like 2A
|
|
chr17_-_21156577
|
0.218
|
NM_152914
|
C17orf103
|
chromosome 17 open reading frame 103
|
|
chr21_+_46494433
|
0.215
|
NM_001112
NM_001160230
NM_015833
NM_015834
|
ADARB1
|
adenosine deaminase, RNA-specific, B1
|
|
chr17_-_65241305
|
0.213
|
NM_014877
|
HELZ
|
helicase with zinc finger
|
|
chr1_+_210502639
|
0.213
|
NM_001170580
|
HHAT
|
hedgehog acyltransferase
|
|
chr13_-_37494370
|
0.213
|
NM_001127217
NM_005905
|
SMAD9
|
SMAD family member 9
|
|
chr12_+_56521953
|
0.186
|
NM_001184796
NM_015292
|
ESYT1
|
extended synaptotagmin-like protein 1
|
|
chr5_+_177540465
|
0.185
|
NM_015111
|
N4BP3
|
NEDD4 binding protein 3
|
|
chr2_-_69870976
|
0.185
|
NM_014911
|
AAK1
|
AP2 associated kinase 1
|
|
chr10_+_102672325
|
0.181
|
NM_001136123
NM_001243770
NM_018121
|
FAM178A
|
family with sequence similarity 178, member A
|
|
chr12_-_96794207
|
0.177
|
NM_001170464
NM_002595
|
CDK17
|
cyclin-dependent kinase 17
|
|
chrX_-_24045302
|
0.173
|
NM_030624
|
KLHL15
|
kelch-like 15 (Drosophila)
|
|
chr16_-_66584045
|
0.169
|
NM_001172643
NM_001172644
NM_001172645
NM_004614
|
TK2
|
thymidine kinase 2, mitochondrial
|
|
chr16_+_85645014
|
0.160
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chr10_+_35535952
|
0.157
|
NM_181698
|
CCNY
|
cyclin Y
|
|
chr1_+_155293727
|
0.157
|
NM_001105205
|
RUSC1
|
RUN and SH3 domain containing 1
|
|
chr1_+_210501595
|
0.150
|
NM_001122834
NM_001170564
|
HHAT
|
hedgehog acyltransferase
|
|
chr20_+_42086468
|
0.147
|
NM_006275
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr10_-_94257470
|
0.143
|
NM_001165946
|
IDE
|
insulin-degrading enzyme
|
|
chr5_+_176560056
|
0.138
|
NM_172349
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr17_-_46178559
|
0.138
|
NM_006807
|
CBX1
|
chromobox homolog 1
|
|
chr17_-_46178761
|
0.138
|
NM_001127228
|
CBX1
|
chromobox homolog 1
|
|
chrX_-_152486073
|
0.138
|
NM_004988
|
MAGEA1
|
melanoma antigen family A, 1 (directs expression of antigen MZ2-E)
|
|
chr12_-_57400199
|
0.137
|
NM_014830
|
ZBTB39
|
zinc finger and BTB domain containing 39
|
|
chr5_+_172483285
|
0.135
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr2_-_218808705
|
0.135
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr1_+_155294217
|
0.135
|
NM_014328
|
RUSC1
|
RUN and SH3 domain containing 1
|
|
chr12_+_105501448
|
0.124
|
NM_015275
|
KIAA1033
|
KIAA1033
|
|
chr20_-_39928708
|
0.118
|
NM_015035
|
ZHX3
|
zinc fingers and homeoboxes 3
|
|
chr22_+_24666749
|
0.118
|
NM_001145468
NM_015330
|
SPECC1L
|
sperm antigen with calponin homology and coiled-coil domains 1-like
|
|
chr10_+_35625608
|
0.112
|
NM_145012
|
CCNY
|
cyclin Y
|
|
chr12_-_110505499
|
0.108
|
NM_207435
|
C12orf76
|
chromosome 12 open reading frame 76
|
|
chr1_+_155290639
|
0.108
|
NM_001105203
NM_001105204
|
RUSC1
|
RUN and SH3 domain containing 1
|
|
chr1_+_154300275
|
0.102
|
NM_020452
|
ATP8B2
|
ATPase, class I, type 8B, member 2
|
|
chr7_+_130794854
|
0.102
|
NM_001145354
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs
|
|
chr1_+_22778343
|
0.102
|
NM_001083621
NM_014870
|
ZBTB40
|
zinc finger and BTB domain containing 40
|
|
chr19_+_47421932
|
0.102
|
NM_004491
|
ARHGAP35
|
Rho GTPase activating protein 35
|
|
chr16_-_75272979
|
0.101
|
NM_001170721
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr16_-_75301950
|
0.101
|
NM_001170715
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr10_+_103113807
|
0.097
|
NM_003939
NM_033637
|
BTRC
|
beta-transducin repeat containing
|
|
chr7_+_100136784
|
0.094
|
NM_006076
|
AGFG2
|
ArfGAP with FG repeats 2
|
|
chr7_+_131012594
|
0.094
|
NM_013255
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs
|
|
chr16_-_75300678
|
0.092
|
NM_001170716
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr14_+_102228134
|
0.091
|
NM_001161725
NM_001161726
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr17_+_61627818
|
0.088
|
NM_005828
|
DCAF7
|
DDB1 and CUL4 associated factor 7
|
|
chr6_-_166075520
|
0.088
|
NM_006661
NM_001130690
|
PDE10A
|
phosphodiesterase 10A
|
|
chr16_-_75282137
|
0.086
|
NM_001170719
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr1_+_26126658
|
0.085
|
NM_020451
NM_206926
|
SEPN1
|
selenoprotein N, 1
|
|
chr16_-_75299713
|
0.083
|
NM_001170720
NM_001170714
NM_001170718
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr2_+_14772809
|
0.083
|
NM_145175
|
FAM84A
|
family with sequence similarity 84, member A
|
|
chr1_+_145507556
|
0.082
|
NM_005105
|
RBM8A
|
RNA binding motif protein 8A
|
|
chr16_+_85646923
|
0.082
|
NM_014615
|
KIAA0182
|
KIAA0182
|
|
chr1_+_245133628
|
0.080
|
NM_001143943
|
EFCAB2
|
EF-hand calcium binding domain 2
|
|
chr15_+_64388082
|
0.079
|
NM_001242933
NM_003099
NM_148955
|
SNX1
|
sorting nexin 1
|
|
chr2_-_20424627
|
0.076
|
NM_002997
|
SDC1
|
syndecan 1
|
|
chr17_+_48796919
|
0.073
|
NM_006107
NM_016424
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae)
|
|
chr16_-_75285383
|
0.071
|
NM_001170717
NM_014567
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr15_-_65579017
|
0.068
|
NM_017851
|
PARP16
|
poly (ADP-ribose) polymerase family, member 16
|
|
chr15_-_60919642
|
0.066
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr12_+_110437233
|
0.063
|
NM_033121
|
ANKRD13A
|
ankyrin repeat domain 13A
|
|
chr18_+_33234532
|
0.058
|
NM_020474
|
GALNT1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1)
|
|
chr11_+_34073188
|
0.058
|
NM_005898
NM_203364
|
CAPRIN1
|
cell cycle associated protein 1
|
|
chr2_-_20425166
|
0.055
|
NM_001006946
|
SDC1
|
syndecan 1
|
|
chr12_+_54393876
|
0.054
|
NM_006897
|
HOXC9
|
homeobox C9
|
|
chr3_-_128902764
|
0.050
|
NM_001127192
NM_001127193
NM_001127194
NM_001127195
NM_001127196
NM_003418
|
CNBP
|
CCHC-type zinc finger, nucleic acid binding protein
|
|
chr2_-_96874569
|
0.048
|
NM_020151
|
STARD7
|
StAR-related lipid transfer (START) domain containing 7
|
|
chr5_-_132299263
|
0.048
|
NM_014423
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr9_-_99329092
|
0.048
|
NM_001077181
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr16_-_69166062
|
0.046
|
NM_001040146
|
CHTF8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae)
|
|
chr20_-_4990913
|
0.045
|
NM_203327
|
SLC23A2
|
solute carrier family 23 (nucleobase transporters), member 2
|
|
chr1_+_6845381
|
0.043
|
NM_001195563
NM_001242701
NM_015215
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr17_+_44668034
|
0.042
|
NM_006178
|
NSF
|
N-ethylmaleimide-sensitive factor
|
|
chr2_+_176957448
|
0.038
|
NM_000523
|
HOXD13
|
homeobox D13
|
|
chr7_+_102073953
|
0.036
|
NM_001126340
NM_032831
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr5_+_176560627
|
0.036
|
NM_022455
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr7_-_143059696
|
0.031
|
NM_001031690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr16_-_69166492
|
0.031
|
NM_001039690
|
CHTF8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae)
|
|
chr12_+_50024326
|
0.030
|
NM_001031698
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae)
|
|
chr10_-_108924287
|
0.029
|
NM_001013031
NM_001206569
NM_001206570
NM_001206571
NM_001206572
NM_052918
|
SORCS1
|
sortilin-related VPS10 domain containing receptor 1
|
|
chr12_+_50017340
|
0.027
|
NM_012272
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae)
|
|
chrX_-_151938194
|
0.023
|
NM_005362
|
MAGEA3
|
melanoma antigen family A, 3
|
|
chr12_+_56137063
|
0.022
|
NM_005811
|
GDF11
|
growth differentiation factor 11
|
|
chr13_+_98795410
|
0.020
|
NM_001001715
NM_005766
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr3_-_128840856
|
0.020
|
NM_001204884
NM_001204883
|
RAB43
|
RAB43, member RAS oncogene family
|
|
chrX_-_54069593
|
0.019
|
NM_001184898
|
PHF8
|
PHD finger protein 8
|
|
chrX_-_131623994
|
0.019
|
NM_001170704
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr4_-_87374282
|
0.018
|
NM_002753
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr22_+_20008602
|
0.017
|
NM_152906
|
C22orf25
|
chromosome 22 open reading frame 25
|
|
chr8_-_101965194
|
0.015
|
NM_001135699
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr15_-_61521478
|
0.013
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr19_+_18682613
|
0.013
|
NM_001033930
NM_003333
|
UBA52
|
ubiquitin A-52 residue ribosomal protein fusion product 1
|
|
chrX_+_151867244
|
0.011
|
NM_005363
NM_175868
|
MAGEA2
MAGEA6
|
melanoma antigen family A, 2
melanoma antigen family A, 6
|
|
chr1_+_111991742
|
0.010
|
NM_001688
|
ATP5F1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit B1
|
|
chr20_+_60718473
|
0.010
|
NM_198935
|
SS18L1
|
synovial sarcoma translocation gene on chromosome 18-like 1
|
|
chr11_+_8703994
|
0.010
|
NM_000990
|
RPL27A
|
ribosomal protein L27a
|
|
chr14_-_50698090
|
0.007
|
NM_006939
|
SOS2
|
son of sevenless homolog 2 (Drosophila)
|
|
chr19_+_797390
|
0.007
|
NM_002819
NM_031990
NM_031991
NM_175847
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chr19_-_9785750
|
0.006
|
NM_001130031
NM_001130032
NM_017656
|
ZNF562
|
zinc finger protein 562
|
|
chr7_+_150725502
|
0.003
|
NM_007188
|
ABCB8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8
|
|
chrX_-_131547536
|
0.003
|
NM_001170701
NM_001170702
NM_001170703
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr22_+_21271619
|
0.003
|
NM_005207
|
CRKL
|
v-crk sarcoma virus CT10 oncogene homolog (avian)-like
|
|
chr18_+_77794345
|
0.003
|
NM_001171967
NM_024805
|
RBFA
|
ribosome binding factor A (putative)
|
|
chr12_-_42538432
|
0.002
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr8_-_30585442
|
0.002
|
NM_000637
NM_001195102
NM_001195103
NM_001195104
|
GSR
|
glutathione reductase
|