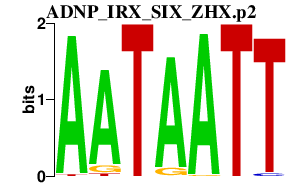
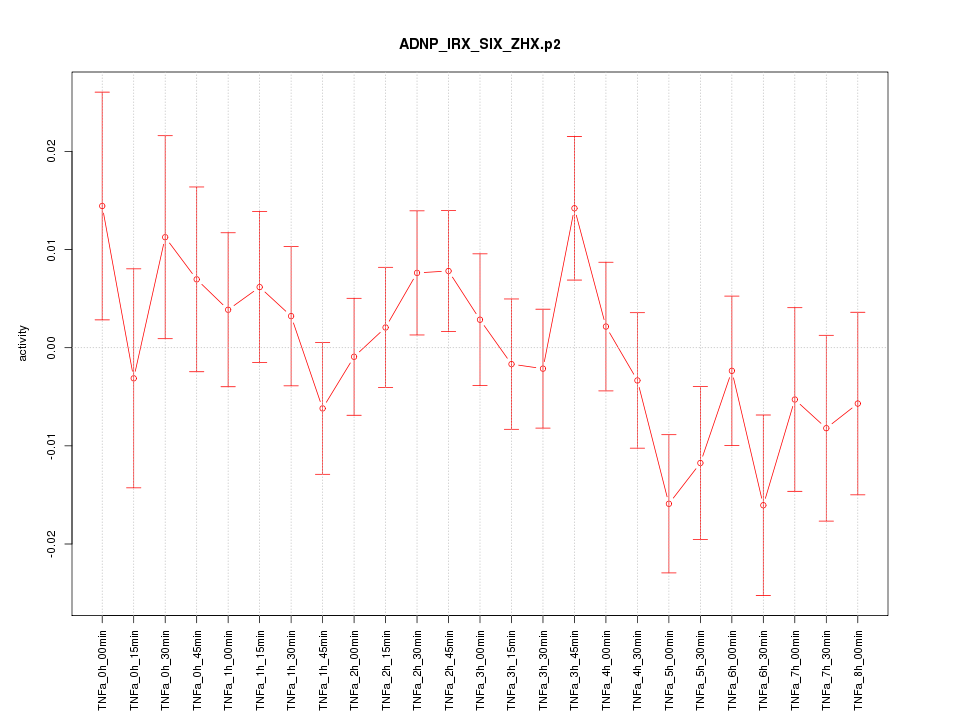
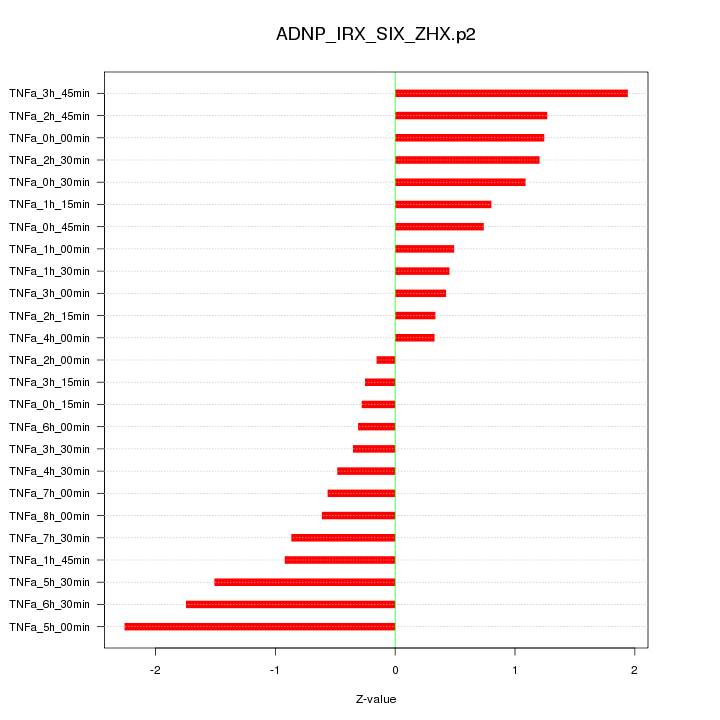
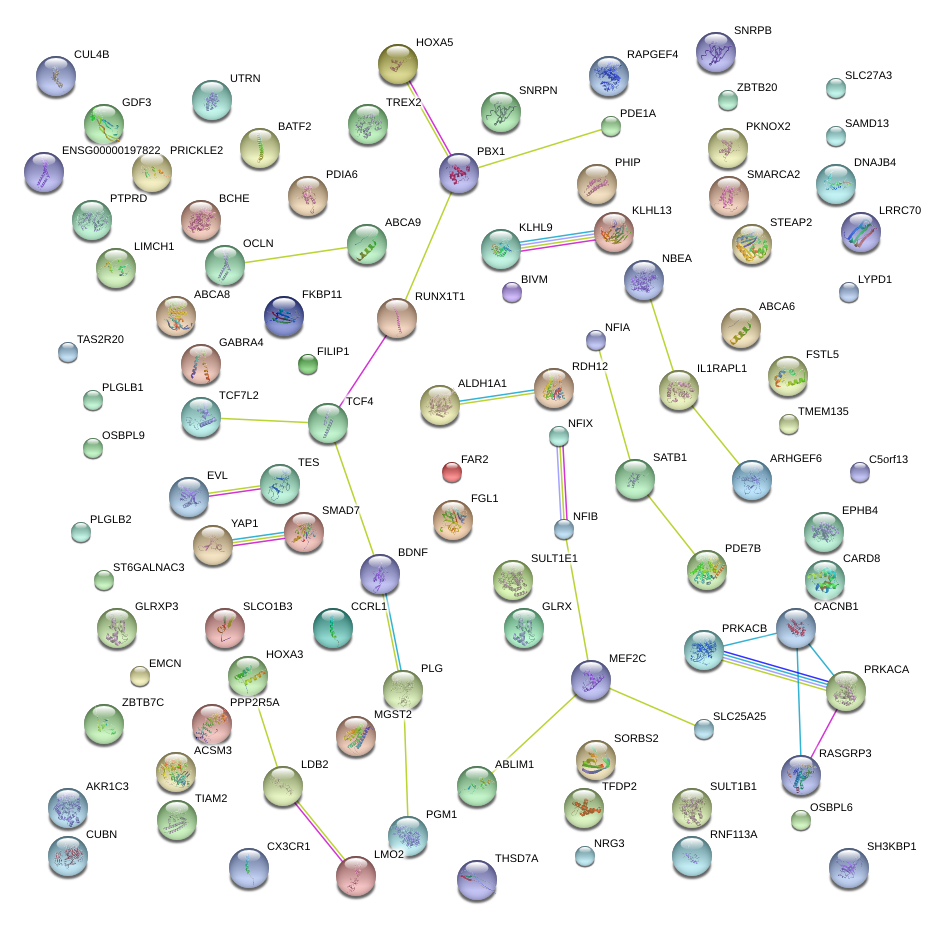
|
chr4_+_41614911
|
7.333
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr4_+_41700736
|
6.490
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr1_+_61869699
|
5.930
|
|
NFIA
|
nuclear factor I/A
|
|
chr9_-_14086901
|
5.890
|
|
NFIB
|
nuclear factor I/B
|
|
chr4_+_41614725
|
5.672
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr1_+_84630014
|
4.834
|
NM_001242857
NM_001242858
NM_001242859
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr4_+_41614933
|
4.377
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr1_+_84630575
|
3.297
|
NM_001242860
NM_001242861
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr1_+_84630374
|
2.739
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr3_+_132316080
|
2.324
|
NM_016557
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr10_+_5136556
|
2.272
|
NM_003739
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr15_+_25281105
|
2.200
|
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr10_+_5136613
|
2.172
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr6_-_76203256
|
2.163
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr1_+_78470593
|
2.156
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr9_+_2015218
|
2.142
|
NM_003070
NM_139045
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2
|
|
chr10_+_5136589
|
2.081
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr11_-_33913835
|
2.044
|
NM_005574
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr12_+_20963637
|
2.022
|
NM_019844
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3
|
|
chr5_-_111312522
|
1.931
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chrX_+_28605558
|
1.920
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr4_+_140587086
|
1.878
|
|
MGST2
|
microsomal glutathione S-transferase 2
|
|
chr19_+_39899749
|
1.874
|
|
|
|
|
chr11_-_71746966
|
1.864
|
|
|
|
|
chr1_+_61547388
|
1.843
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr4_-_101439071
|
1.839
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr9_-_75567915
|
1.774
|
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr7_+_115863004
|
1.666
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr2_+_173686314
|
1.657
|
NM_001100397
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr3_-_18480203
|
1.634
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr4_-_16900028
|
1.588
|
|
LDB2
|
LIM domain binding 2
|
|
chr11_+_125034558
|
1.552
|
NM_022062
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr3_-_64211111
|
1.534
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr4_-_70626429
|
1.455
|
NM_014465
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1
|
|
chr11_+_87032428
|
1.411
|
|
TMEM135
|
transmembrane protein 135
|
|
chr3_-_114478117
|
1.377
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr4_-_16900348
|
1.348
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr4_-_70725792
|
1.335
|
NM_005420
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1
|
|
chr8_-_93075190
|
1.328
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr7_-_27183225
|
1.295
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr2_-_183387063
|
1.270
|
NM_005019
NM_001003683
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr18_-_53089722
|
1.261
|
NM_001243233
|
TCF4
|
transcription factor 4
|
|
chr1_+_84647342
|
1.244
|
NM_001242862
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr4_-_186577858
|
1.244
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chrX_-_119709618
|
1.240
|
NM_003588
|
CUL4B
|
cullin 4B
|
|
chr7_+_89843133
|
1.229
|
NM_001040666
|
STEAP2
|
STEAP family member 2, metalloreductase
|
|
chr8_-_17752847
|
1.228
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr1_+_84768333
|
1.212
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr14_+_100563817
|
1.190
|
|
EVL
|
Enah/Vasp-like
|
|
chr1_+_76540374
|
1.188
|
NM_001160011
NM_152996
|
ST6GALNAC3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3
|
|
chr18_-_46469118
|
1.183
|
NM_001190823
|
SMAD7
|
SMAD family member 7
|
|
chr17_-_67138013
|
1.179
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chrX_-_19817854
|
1.172
|
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr1_+_212475147
|
1.130
|
NM_001199756
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha
|
|
chr5_-_88199868
|
1.117
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr4_-_186732208
|
1.095
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_+_84767288
|
1.077
|
NM_001134663
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chrX_-_135863104
|
1.071
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr4_-_163085117
|
1.056
|
NM_001128427
NM_001128428
NM_020116
|
FSTL5
|
follistatin-like 5
|
|
chrX_-_117119282
|
1.047
|
NM_001168299
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr9_-_8733893
|
1.012
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr10_+_83637442
|
1.011
|
NM_001165973
|
NRG3
|
neuregulin 3
|
|
chr8_-_93107442
|
0.992
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr7_-_11871735
|
0.979
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chrX_-_135863033
|
0.977
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr8_-_93111915
|
0.976
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr4_+_140586921
|
0.966
|
NM_001204366
NM_001204367
NM_001204368
NM_002413
|
MGST2
|
microsomal glutathione S-transferase 2
|
|
chr2_-_87248942
|
0.952
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr6_+_155538096
|
0.941
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr5_+_68800003
|
0.930
|
NM_001205255
|
OCLN
|
occludin
|
|
chr6_+_136172802
|
0.928
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr11_-_27742224
|
0.926
|
NM_001143805
NM_001143806
NM_170732
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr5_+_61874561
|
0.924
|
NM_181506
|
LRRC70
|
leucine rich repeat containing 70
|
|
chr9_-_14398981
|
0.922
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr1_+_153747767
|
0.895
|
NM_024330
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3
|
|
chr6_+_144612872
|
0.895
|
NM_007124
|
UTRN
|
utrophin
|
|
chr3_+_132318980
|
0.882
|
NM_178445
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr1_+_164818083
|
0.879
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr3_-_141747434
|
0.878
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr16_+_20775311
|
0.870
|
NM_005622
NM_202000
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3
|
|
chr5_-_111091947
|
0.868
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr2_+_179184970
|
0.868
|
NM_145739
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr2_+_200323088
|
0.837
|
|
|
|
|
chr2_-_133429069
|
0.835
|
NM_001077427
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr7_-_100425035
|
0.831
|
NM_004444
|
EPHB4
|
EPH receptor B4
|
|
chr13_+_103459495
|
0.825
|
NM_001204425
|
BIVM-ERCC5
BIVM
|
BIVM-ERCC5 readthrough
basic, immunoglobulin-like variable motif containing
|
|
chr10_-_17171789
|
0.824
|
NM_001081
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor)
|
|
chr6_-_79675672
|
0.815
|
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr12_-_7848324
|
0.814
|
NM_020634
|
GDF3
|
growth differentiation factor 3
|
|
chr1_+_52082545
|
0.806
|
NM_024586
NM_148906
NM_148908
NM_148909
|
OSBPL9
|
oxysterol binding protein-like 9
|
|
chr17_-_66951381
|
0.802
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr8_-_93029864
|
0.802
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr14_+_68168602
|
0.796
|
NM_152443
|
RDH12
|
retinol dehydrogenase 12 (all-trans/9-cis/11-cis)
|
|
chr3_-_18466759
|
0.795
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr3_-_165555167
|
0.791
|
|
BCHE
|
butyrylcholinesterase
|
|
chr9_+_130860816
|
0.781
|
NM_052901
NM_001006643
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr19_-_48752921
|
0.781
|
NM_001184901
NM_001184903
NM_014959
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr9_+_130860769
|
0.778
|
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr7_-_27166638
|
0.762
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr13_+_36050885
|
0.755
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr18_-_45663667
|
0.751
|
NM_001039360
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr17_-_67057046
|
0.750
|
NM_080283
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9
|
|
chr8_-_93115453
|
0.749
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr12_+_29376596
|
0.747
|
NM_018099
|
FAR2
|
fatty acyl CoA reductase 2
|
|
chr2_-_10942642
|
0.739
|
|
PDIA6
|
protein disulfide isomerase family A, member 6
|
|
chr11_+_101983170
|
0.736
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr10_-_116392099
|
0.728
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr1_+_64088886
|
0.726
|
NM_001172818
|
PGM1
|
phosphoglucomutase 1
|
|
chr2_+_33738941
|
0.724
|
NM_015376
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr5_-_95158465
|
0.717
|
NM_001118890
NM_001243658
NM_001243659
NM_002064
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr5_-_168271852
|
0.711
|
|
|
|
|
chr2_+_88047603
|
0.702
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr4_-_46996423
|
0.694
|
NM_000809
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4
|
|
chr12_-_11150473
|
0.692
|
NM_176889
|
TAS2R20
|
taste receptor, type 2, member 20
|
|
chr3_+_25470067
|
0.687
|
|
RARB
|
retinoic acid receptor, beta
|
|
chr7_+_80275634
|
0.684
|
NM_001127443
NM_001127444
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr21_-_39870303
|
0.676
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr13_+_53035227
|
0.675
|
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr7_-_86595203
|
0.673
|
NM_152748
|
KIAA1324L
|
KIAA1324-like
|
|
chr3_-_69037498
|
0.672
|
|
C3orf64
|
chromosome 3 open reading frame 64
|
|
chr16_+_82068830
|
0.666
|
NM_002153
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr11_+_92085261
|
0.663
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr8_-_6420454
|
0.660
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr5_-_39394423
|
0.658
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr9_+_120466607
|
0.655
|
|
TLR4
|
toll-like receptor 4
|
|
chr5_-_95158365
|
0.652
|
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr6_+_132873829
|
0.650
|
NM_053278
|
TAAR8
|
trace amine associated receptor 8
|
|
chr16_+_82068958
|
0.647
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr11_-_27722599
|
0.645
|
NM_001143808
NM_001143809
NM_001143810
NM_001143811
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr16_+_82068910
|
0.641
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chrX_-_19817907
|
0.641
|
NM_001024666
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr15_+_57511616
|
0.639
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr12_-_10021932
|
0.638
|
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr17_+_3377346
|
0.635
|
NM_001128085
|
ASPA
|
aspartoacylase
|
|
chr12_+_14538232
|
0.632
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr2_-_44586854
|
0.626
|
NM_001042385
NM_001042386
|
PREPL
|
prolyl endopeptidase-like
|
|
chr9_-_124989755
|
0.622
|
NM_001242334
|
LHX6
|
LIM homeobox 6
|
|
chr3_-_128294928
|
0.619
|
NM_007354
|
C3orf27
|
chromosome 3 open reading frame 27
|
|
chr4_-_186733372
|
0.614
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr17_-_46651809
|
0.614
|
NM_002146
|
HOXB3
|
homeobox B3
|
|
chr18_-_53303129
|
0.607
|
NM_001243226
|
TCF4
|
transcription factor 4
|
|
chr10_+_5005685
|
0.604
|
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr14_-_38725253
|
0.601
|
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr12_+_93963597
|
0.596
|
NM_003877
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr2_-_211018294
|
0.595
|
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr1_-_197552304
|
0.592
|
|
DENND1B
|
DENN/MADD domain containing 1B
|
|
chr16_+_8806825
|
0.590
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr7_-_26232148
|
0.590
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr20_-_34117447
|
0.583
|
NM_001145350
|
C20orf173
|
chromosome 20 open reading frame 173
|
|
chr5_+_73109342
|
0.577
|
NM_001244364
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr1_-_110933635
|
0.572
|
|
SLC16A4
|
solute carrier family 16, member 4 (monocarboxylic acid transporter 5)
|
|
chr10_+_35426709
|
0.571
|
NM_183011
NM_183012
|
CREM
|
cAMP responsive element modulator
|
|
chrX_+_85517970
|
0.571
|
NM_001139515
|
DACH2
|
dachshund homolog 2 (Drosophila)
|
|
chr8_-_74659942
|
0.569
|
NM_001164384
|
STAU2
|
staufen, RNA binding protein, homolog 2 (Drosophila)
|
|
chr3_-_165555252
|
0.569
|
NM_000055
|
BCHE
|
butyrylcholinesterase
|
|
chr10_-_126716452
|
0.562
|
NM_022802
|
CTBP2
|
C-terminal binding protein 2
|
|
chr2_+_189839132
|
0.562
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr1_-_110933683
|
0.561
|
NM_001201546
NM_001201547
NM_001201548
NM_001201549
NM_004696
|
SLC16A4
|
solute carrier family 16, member 4 (monocarboxylic acid transporter 5)
|
|
chrX_+_79675945
|
0.561
|
NM_152630
|
FAM46D
|
family with sequence similarity 46, member D
|
|
chr5_+_140602914
|
0.553
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr12_-_14132920
|
0.551
|
NM_000834
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B
|
|
chr6_-_89927495
|
0.544
|
NM_002042
|
GABRR1
|
gamma-aminobutyric acid (GABA) receptor, rho 1
|
|
chr10_-_116286570
|
0.542
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr8_-_6837568
|
0.540
|
NM_004084
NM_001042500
|
DEFA1
DEFA1B
|
defensin, alpha 1
defensin, alpha 1B
|
|
chr21_-_40033617
|
0.540
|
NM_001243432
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr8_-_93107643
|
0.539
|
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr21_+_38791206
|
0.536
|
NM_130436
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chrX_-_117107700
|
0.533
|
NM_033495
NM_001168300
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chrX_+_135230736
|
0.532
|
NM_001159700
|
FHL1
|
four and a half LIM domains 1
|
|
chr15_-_37391893
|
0.530
|
|
MEIS2
|
Meis homeobox 2
|
|
chr3_-_137851123
|
0.529
|
NM_016161
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase
|
|
chr4_-_138453628
|
0.525
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr18_+_29769986
|
0.521
|
NM_005925
|
MEP1B
|
meprin A, beta
|
|
chr10_+_102123420
|
0.521
|
|
|
|
|
chr7_-_138458781
|
0.520
|
NM_130841
|
ATP6V0A4
|
ATPase, H+ transporting, lysosomal V0 subunit a4
|
|
chr8_-_6875777
|
0.520
|
NM_004084
NM_005217
NM_001042500
|
DEFA1
DEFA3
DEFA1B
|
defensin, alpha 1
defensin, alpha 3, neutrophil-specific
defensin, alpha 1B
|
|
chr5_+_95066628
|
0.520
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|
|
chr8_-_72268720
|
0.518
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr3_-_168864066
|
0.513
|
NM_001105077
NM_001163999
NM_005241
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chrX_+_66788682
|
0.513
|
NM_001011645
|
AR
|
androgen receptor
|
|
chr5_-_42811959
|
0.512
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chrX_+_56755666
|
0.512
|
|
LOC550643
|
uncharacterized LOC550643
|
|
chr10_-_116286661
|
0.500
|
NM_006720
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr12_+_101988746
|
0.500
|
NM_002465
NM_206819
NM_206820
NM_206821
|
MYBPC1
|
myosin binding protein C, slow type
|
|
chr11_+_114167197
|
0.499
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr11_+_114167168
|
0.499
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr1_-_242612757
|
0.495
|
NM_001195811
NM_001195812
|
PLD5
|
phospholipase D family, member 5
|
|
chr18_-_53177999
|
0.495
|
NM_001243231
|
TCF4
|
transcription factor 4
|
|
chr14_-_90085325
|
0.495
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr3_-_112359251
|
0.490
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr7_-_27205123
|
0.487
|
|
HOXA9
|
homeobox A9
|
|
chr1_+_228645807
|
0.487
|
NM_175055
|
HIST3H2BB
|
histone cluster 3, H2bb
|
|
chr1_+_32043745
|
0.487
|
NM_001204415
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr10_-_50732142
|
0.484
|
NM_170753
|
PGBD3
|
piggyBac transposable element derived 3
|
|
chr10_+_124320180
|
0.481
|
NM_004406
NM_007329
NM_017579
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr10_+_5005601
|
0.480
|
|
AKR1C1
AKR1C3
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chrX_-_18690187
|
0.478
|
NM_000330
|
RS1
|
retinoschisin 1
|
|
chr13_-_45010937
|
0.477
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr13_-_45048436
|
0.476
|
NM_001243798
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr4_-_79103271
|
0.473
|
|
|
|