|
chr20_+_58515408
|
2.784
|
NM_001190827
NM_022106
|
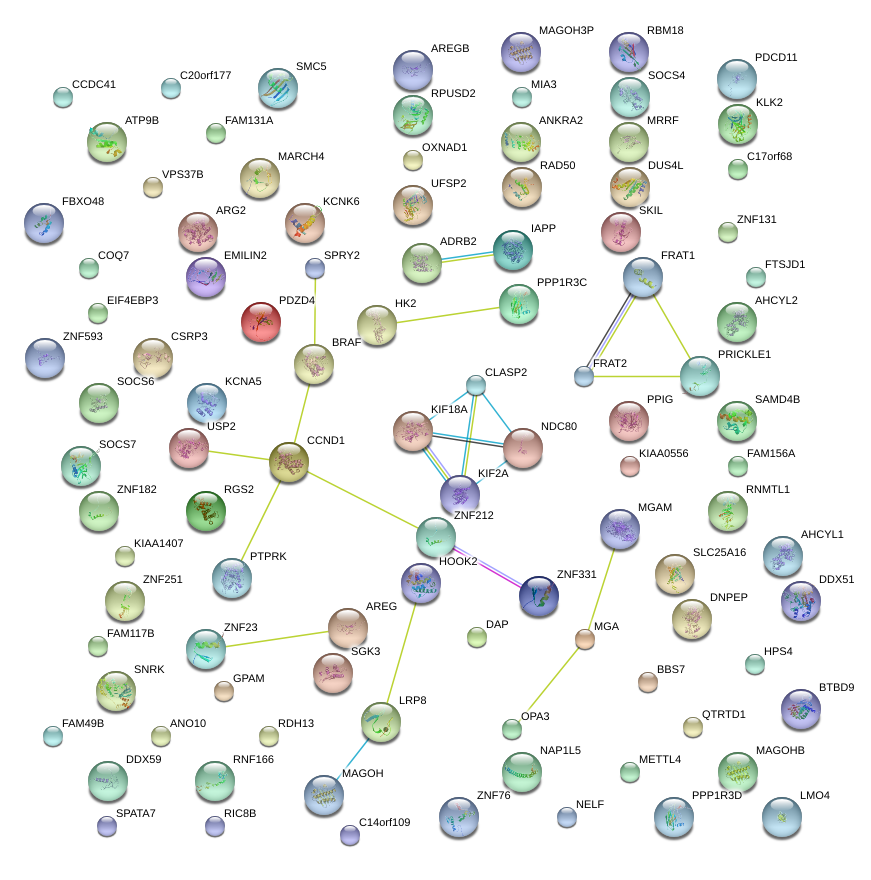
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr3_+_170075449
|
2.269
|
NM_001145098
NM_005414
|
SKIL
|
SKI-like oncogene
|
|
chr4_-_89618928
|
2.233
|
NM_153757
|
NAP1L5
|
nucleosome assembly protein 1-like 5
|
|
chr10_-_93392857
|
2.120
|
NM_005398
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr9_-_125027017
|
2.040
|
|
RBM18
|
RNA binding motif protein 18
|
|
chr13_-_80912971
|
2.039
|
|
SPRY2
|
sprouty homolog 2 (Drosophila)
|
|
chr16_-_88772732
|
1.967
|
NM_001171815
|
RNF166
|
ring finger protein 166
|
|
chr16_-_88772807
|
1.889
|
NM_178841
|
RNF166
|
ring finger protein 166
|
|
chr9_-_125027080
|
1.861
|
NM_033117
|
RBM18
|
RNA binding motif protein 18
|
|
chr11_+_69455938
|
1.749
|
|
CCND1
|
cyclin D1
|
|
chr19_+_54058493
|
1.715
|
NM_001079907
|
ZNF331
|
zinc finger protein 331
|
|
chr3_+_170075494
|
1.646
|
|
SKIL
|
SKI-like oncogene
|
|
chr11_-_119252267
|
1.599
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr12_-_10766130
|
1.589
|
NM_018048
|
MAGOHB
|
mago-nashi homolog B (Drosophila)
|
|
chr10_-_99094427
|
1.544
|
NM_012083
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2
|
|
chr14_+_68086570
|
1.543
|
NM_001172
|
ARG2
|
arginase, type II
|
|
chr19_+_38810540
|
1.486
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr12_-_123380628
|
1.481
|
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr3_+_16306666
|
1.478
|
NM_138381
|
OXNAD1
|
oxidoreductase NAD-binding domain containing 1
|
|
chr19_-_46088084
|
1.464
|
NM_001017989
NM_025136
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr12_+_107168336
|
1.463
|
NM_018157
|
RIC8B
|
resistance to inhibitors of cholinesterase 8 homolog B (C. elegans)
|
|
chr12_-_123380682
|
1.443
|
NM_024667
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr20_-_58515258
|
1.426
|
NM_006242
|
PPP1R3D
|
protein phosphatase 1, regulatory subunit 3D
|
|
chr6_-_15248886
|
1.348
|
|
|
|
|
chr6_-_38607635
|
1.339
|
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chr12_-_42877413
|
1.333
|
NM_001144881
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chr15_+_40861491
|
1.314
|
|
RPUSD2
|
RNA pseudouridylate synthase domain containing 2
|
|
chr15_+_40861509
|
1.309
|
|
RPUSD2
|
RNA pseudouridylate synthase domain containing 2
|
|
chr15_+_40861531
|
1.297
|
NM_152260
|
RPUSD2
|
RNA pseudouridylate synthase domain containing 2
|
|
chr9_+_125027152
|
1.295
|
|
MRRF
|
mitochondrial ribosome recycling factor
|
|
chr3_+_184053682
|
1.282
|
NM_001171093
|
FAM131A
|
family with sequence similarity 131, member A
|
|
chr14_+_93651295
|
1.260
|
NM_015676
NM_001098621
|
C14orf109
|
chromosome 14 open reading frame 109
|
|
chr15_+_40861527
|
1.254
|
|
RPUSD2
|
RNA pseudouridylate synthase domain containing 2
|
|
chr1_-_53793743
|
1.233
|
NM_001018054
NM_004631
NM_017522
NM_033300
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor
|
|
chr18_-_2571422
|
1.216
|
NM_022840
|
METTL4
|
methyltransferase like 4
|
|
chr12_-_94853453
|
1.214
|
|
CCDC41
|
coiled-coil domain containing 41
|
|
chr2_+_75061234
|
1.207
|
|
HK2
|
hexokinase 2
|
|
chr22_-_26879724
|
1.206
|
|
HPS4
|
Hermansky-Pudlak syndrome 4
|
|
chr15_+_41952641
|
1.197
|
|
MGA
|
MAX gene associated
|
|
chr16_+_19078958
|
1.189
|
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr16_-_71496088
|
1.188
|
NM_145911
|
ZNF23
|
zinc finger protein 23 (KOX 16)
|
|
chr11_+_28129797
|
1.172
|
NM_001113528
NM_152636
|
METTL15
|
methyltransferase like 15
|
|
chr17_+_36507708
|
1.153
|
NM_014598
|
SOCS7
|
suppressor of cytokine signaling 7
|
|
chr19_-_55574434
|
1.144
|
NM_001145971
|
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis)
|
|
chr18_+_2571587
|
1.142
|
|
NDC80
|
NDC80 kinetochore complex component homolog (S. cerevisiae)
|
|
chr18_+_2571509
|
1.137
|
NM_006101
|
NDC80
|
NDC80 kinetochore complex component homolog (S. cerevisiae)
|
|
chr1_+_222791151
|
1.105
|
NM_198551
|
MIA3
|
melanoma inhibitory activity family, member 3
|
|
chr6_-_38607923
|
1.092
|
NM_001099272
NM_052893
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chr7_-_140624280
|
1.073
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr3_-_113775363
|
1.073
|
NM_020817
|
KIAA1407
|
KIAA1407
|
|
chr10_-_113943467
|
1.054
|
NM_001244949
NM_020918
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr11_-_28129646
|
1.050
|
|
KIF18A
|
kinesin family member 18A
|
|
chr17_+_685606
|
1.041
|
|
RNMTL1
|
RNA methyltransferase like 1
|
|
chr1_+_26496382
|
1.022
|
NM_015871
|
ZNF593
|
zinc finger protein 593
|
|
chr3_-_33759684
|
1.022
|
NM_015097
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr3_+_183165402
|
1.014
|
|
LOC100505687
|
uncharacterized LOC100505687
|
|
chr5_-_72861376
|
0.986
|
NM_023039
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2
|
|
chr8_+_67624941
|
0.984
|
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3
|
|
chr11_+_69455837
|
0.978
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr2_+_203499819
|
0.968
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr1_+_110527327
|
0.962
|
NM_001242673
NM_006621
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr17_+_685512
|
0.948
|
NM_018146
|
RNMTL1
|
RNA methyltransferase like 1
|
|
chr3_+_170075521
|
0.939
|
|
SKIL
|
SKI-like oncogene
|
|
chr10_+_99079017
|
0.929
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr2_-_68694389
|
0.926
|
NM_001024680
|
FBXO48
|
F-box protein 48
|
|
chr2_-_217236666
|
0.925
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr21_-_30365111
|
0.925
|
NM_015565
|
LTN1
|
listerin E3 ubiquitin protein ligase 1
|
|
chr18_+_2906956
|
0.914
|
|
EMILIN2
|
elastin microfibril interfacer 2
|
|
chr1_-_200638996
|
0.903
|
|
DDX59
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 59
|
|
chr11_-_65337743
|
0.898
|
|
LOC254100
|
uncharacterized LOC254100
|
|
chr1_+_87794576
|
0.897
|
|
LMO4
|
LIM domain only 4
|
|
chr5_-_10761344
|
0.893
|
NM_004394
|
DAP
|
death-associated protein
|
|
chr19_+_39833096
|
0.891
|
|
SAMD4B
|
sterile alpha motif domain containing 4B
|
|
chr9_+_125026881
|
0.888
|
NM_001173512
NM_138777
|
MRRF
|
mitochondrial ribosome recycling factor
|
|
chr10_-_70287249
|
0.881
|
NM_152707
|
SLC25A16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16
|
|
chr5_+_43121641
|
0.879
|
NM_003432
|
ZNF131
|
zinc finger protein 131
|
|
chr19_+_38810751
|
0.879
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr12_-_94853696
|
0.875
|
NM_001042399
NM_016122
|
CCDC41
|
coiled-coil domain containing 41
|
|
chrX_+_52926317
|
0.861
|
NM_001242491
|
FAM156A
|
family with sequence similarity 156, member A
|
|
chr17_-_8151353
|
0.853
|
|
CTC1
|
CTS telomere maintenance complex component 1
|
|
chr5_-_10761341
|
0.852
|
|
DAP
|
death-associated protein
|
|
chr6_+_35227234
|
0.843
|
|
ZNF76
|
zinc finger protein 76
|
|
chrX_+_52926334
|
0.836
|
|
FAM156A
FAM156B
|
family with sequence similarity 156, member A
family with sequence similarity 156, member B
|
|
chr4_-_186347037
|
0.829
|
NM_018359
|
UFSP2
|
UFM1-specific peptidase 2
|
|
chr8_-_145980955
|
0.820
|
NM_138367
|
ZNF251
|
zinc finger protein 251
|
|
chr15_+_41952572
|
0.807
|
NM_001080541
NM_001164273
|
MGA
|
MAX gene associated
|
|
chr6_-_128841432
|
0.806
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr2_+_170441102
|
0.803
|
|
PPIG
|
peptidylprolyl isomerase G (cyclophilin G)
|
|
chr9_+_72873820
|
0.795
|
NM_015110
|
SMC5
|
structural maintenance of chromosomes 5
|
|
chr12_+_58166341
|
0.786
|
NM_015433
NM_206914
|
METTL21B
|
methyltransferase like 21B
|
|
chr4_+_16228273
|
0.782
|
|
FLJ39653
|
uncharacterized FLJ39653
|
|
chr8_-_131028674
|
0.782
|
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr22_-_26879761
|
0.782
|
NM_022081
|
HPS4
|
Hermansky-Pudlak syndrome 4
|
|
chr3_+_43328001
|
0.781
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr9_-_140353785
|
0.780
|
NM_001130969
NM_001130970
NM_001130971
NM_001178064
NM_015537
|
NELF
|
nasal embryonic LHRH factor
|
|
chr3_+_113775658
|
0.778
|
|
QTRTD1
|
queuine tRNA-ribosyltransferase domain containing 1
|
|
chr4_-_122791468
|
0.777
|
NM_018190
NM_176824
|
BBS7
|
Bardet-Biedl syndrome 7
|
|
chrX_-_47863260
|
0.776
|
NM_001007088
NM_006962
NM_001178099
|
ZNF182
|
zinc finger protein 182
|
|
chr12_+_132629009
|
0.769
|
|
|
|
|
chr3_+_113775580
|
0.769
|
NM_024638
|
QTRTD1
|
queuine tRNA-ribosyltransferase domain containing 1
|
|
chr1_+_192778168
|
0.765
|
NM_002923
|
RGS2
|
regulator of G-protein signaling 2, 24kDa
|
|
chr5_+_148206155
|
0.762
|
NM_000024
|
ADRB2
|
adrenergic, beta-2-, receptor, surface
|
|
chr14_+_88851987
|
0.762
|
NM_001040428
NM_018418
|
SPATA7
|
spermatogenesis associated 7
|
|
chr12_-_132628841
|
0.758
|
NM_175066
|
DDX51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51
|
|
chr10_-_93392744
|
0.758
|
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr16_-_71323090
|
0.756
|
NM_001099642
NM_018348
|
FTSJD1
|
FtsJ methyltransferase domain containing 1
|
|
chr5_+_139927182
|
0.755
|
NM_003732
|
EIF4EBP3
|
eukaryotic translation initiation factor 4E binding protein 3
|
|
chr8_-_131028646
|
0.753
|
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr19_+_54057898
|
0.752
|
NM_001253801
|
ZNF331
|
zinc finger protein 331
|
|
chr16_+_27561441
|
0.747
|
NM_015202
|
KIAA0556
|
KIAA0556
|
|
chr2_+_170440846
|
0.736
|
NM_004792
|
PPIG
|
peptidylprolyl isomerase G (cyclophilin G)
|
|
chr5_+_131892556
|
0.734
|
NM_005732
|
RAD50
|
RAD50 homolog (S. cerevisiae)
|
|
chr18_+_76829410
|
0.723
|
|
ATP9B
|
ATPase, class II, type 9B
|
|
chr1_-_53793583
|
0.722
|
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor
|
|
chr7_+_107204402
|
0.721
|
NM_181581
|
DUS4L
|
dihydrouridine synthase 4-like (S. cerevisiae)
|
|
chr4_+_75310852
|
0.721
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr9_-_140353748
|
0.719
|
|
NELF
|
nasal embryonic LHRH factor
|
|
chr19_-_49496520
|
0.716
|
|
GYS1
|
glycogen synthase 1 (muscle)
|
|
chrX_+_52926358
|
0.715
|
|
FAM156B
|
family with sequence similarity 156, member B
|
|
chr4_-_119757183
|
0.715
|
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr7_+_100027253
|
0.713
|
NM_019606
|
MEPCE
|
methylphosphate capping enzyme
|
|
chr19_-_49496369
|
0.712
|
|
GYS1
|
glycogen synthase 1 (muscle)
|
|
chr5_-_10761179
|
0.710
|
|
DAP
|
death-associated protein
|
|
chr4_+_113558119
|
0.709
|
NM_016648
|
LARP7
|
La ribonucleoprotein domain family, member 7
|
|
chr16_-_3767449
|
0.705
|
NM_016292
|
TRAP1
|
TNF receptor-associated protein 1
|
|
chr13_+_49822046
|
0.701
|
NM_001193478
NM_030911
|
CDADC1
|
cytidine and dCMP deaminase domain containing 1
|
|
chr4_-_146019296
|
0.698
|
NM_014885
|
ANAPC10
|
anaphase promoting complex subunit 10
|
|
chr3_-_141944441
|
0.696
|
NM_001039547
|
GK5
|
glycerol kinase 5 (putative)
|
|
chr19_-_36036390
|
0.695
|
|
LOC100506469
|
uncharacterized LOC100506469
|
|
chr4_-_79860369
|
0.686
|
NM_001040202
|
PAQR3
|
progestin and adipoQ receptor family member III
|
|
chr5_-_133304274
|
0.686
|
NM_020199
|
C5orf15
|
chromosome 5 open reading frame 15
|
|
chr1_-_200639091
|
0.685
|
NM_001031725
|
DDX59
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 59
|
|
chr2_-_202645894
|
0.683
|
NM_001135745
NM_020919
|
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile)
|
|
chr10_+_23728197
|
0.682
|
NM_001145373
|
OTUD1
|
OTU domain containing 1
|
|
chr13_+_51483813
|
0.681
|
NM_001142279
NM_024570
|
RNASEH2B
|
ribonuclease H2, subunit B
|
|
chr5_+_41904459
|
0.675
|
NM_175921
|
C5orf51
|
chromosome 5 open reading frame 51
|
|
chr16_+_2802683
|
0.671
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr8_-_21966812
|
0.669
|
NM_024815
|
NUDT18
|
nudix (nucleoside diphosphate linked moiety X)-type motif 18
|
|
chr12_+_49372235
|
0.667
|
NM_005430
|
WNT1
|
wingless-type MMTV integration site family, member 1
|
|
chr14_+_88852065
|
0.667
|
|
SPATA7
|
spermatogenesis associated 7
|
|
chr15_+_55700722
|
0.666
|
NM_001198784
|
FLJ27352
|
uncharacterized LOC145788
|
|
chr11_-_77184549
|
0.664
|
NM_001128620
NM_002576
|
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1
|
|
chr19_+_58695199
|
0.664
|
|
ZNF274
|
zinc finger protein 274
|
|
chr17_-_72869068
|
0.661
|
NM_004110
NM_024417
|
FDXR
|
ferredoxin reductase
|
|
chr9_+_140083053
|
0.661
|
NM_003731
|
SSNA1
|
Sjogren syndrome nuclear autoantigen 1
|
|
chr10_+_102759607
|
0.660
|
|
LZTS2
|
leucine zipper, putative tumor suppressor 2
|
|
chr7_-_35734728
|
0.660
|
|
HERPUD2
|
HERPUD family member 2
|
|
chr6_+_49431090
|
0.659
|
NM_018132
|
CENPQ
|
centromere protein Q
|
|
chr3_+_44771097
|
0.659
|
NM_145044
|
ZNF501
|
zinc finger protein 501
|
|
chr16_+_19078916
|
0.657
|
NM_016138
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr10_-_38265452
|
0.655
|
NM_145011
|
ZNF25
|
zinc finger protein 25
|
|
chr7_+_116660263
|
0.653
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr19_-_49496566
|
0.650
|
NM_001161587
NM_002103
|
GYS1
|
glycogen synthase 1 (muscle)
|
|
chr9_+_111696426
|
0.645
|
|
FAM206A
|
family with sequence similarity 206, member A
|
|
chr11_+_72525352
|
0.645
|
NM_033388
|
ATG16L2
|
ATG16 autophagy related 16-like 2 (S. cerevisiae)
|
|
chr7_-_35734472
|
0.644
|
|
HERPUD2
|
HERPUD family member 2
|
|
chr14_-_77494974
|
0.637
|
NM_024496
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like
|
|
chr11_-_28129701
|
0.636
|
NM_031217
|
KIF18A
|
kinesin family member 18A
|
|
chrX_-_52987610
|
0.635
|
NM_001242491
|
FAM156A
FAM156B
|
family with sequence similarity 156, member A
family with sequence similarity 156, member B
|
|
chr2_+_220144024
|
0.634
|
NM_001039550
NM_006736
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2
|
|
chr4_-_148605135
|
0.631
|
NM_138364
|
PRMT10
|
protein arginine methyltransferase 10 (putative)
|
|
chr20_+_23342786
|
0.629
|
|
GZF1
|
GDNF-inducible zinc finger protein 1
|
|
chr9_+_111696672
|
0.623
|
NM_017832
|
FAM206A
|
family with sequence similarity 206, member A
|
|
chr13_+_45915479
|
0.619
|
|
LOC100190939
|
uncharacterized LOC100190939
|
|
chr1_+_162760491
|
0.614
|
NM_016371
|
HSD17B7
|
hydroxysteroid (17-beta) dehydrogenase 7
|
|
chr17_+_74722892
|
0.614
|
NM_001080510
NM_001206983
NM_001206984
NM_001206985
NM_001206986
NM_001206987
|
METTL23
|
methyltransferase like 23
|
|
chr5_-_54603401
|
0.613
|
NM_019030
|
DHX29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29
|
|
chr2_-_25194772
|
0.612
|
NM_001198559
NM_016544
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27
|
|
chr10_+_121578768
|
0.611
|
NM_001243194
|
INPP5F
|
inositol polyphosphate-5-phosphatase F
|
|
chr14_+_68086655
|
0.611
|
|
ARG2
|
arginase, type II
|
|
chr16_-_86588680
|
0.610
|
NM_001159380
NM_001159377
NM_001159378
NM_001159379
NM_022764
|
MTHFSD
|
methenyltetrahydrofolate synthetase domain containing
|
|
chr16_+_2802640
|
0.607
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr19_-_47617008
|
0.602
|
NM_015168
|
ZC3H4
|
zinc finger CCCH-type containing 4
|
|
chr14_+_60558628
|
0.592
|
NM_022495
|
C14orf135
|
chromosome 14 open reading frame 135
|
|
chr1_-_23495284
|
0.591
|
|
LUZP1
|
leucine zipper protein 1
|
|
chr2_-_64371444
|
0.589
|
NM_020651
|
PELI1
|
pellino homolog 1 (Drosophila)
|
|
chr1_-_23495331
|
0.589
|
|
LUZP1
|
leucine zipper protein 1
|
|
chr15_+_49447951
|
0.587
|
NM_001001556
|
GALK2
|
galactokinase 2
|
|
chr15_+_50716568
|
0.586
|
NM_001128610
NM_001128611
NM_005154
|
USP8
|
ubiquitin specific peptidase 8
|
|
chr9_-_139981102
|
0.583
|
|
LOC100289341
|
uncharacterized LOC100289341
|
|
chr11_+_9482511
|
0.582
|
NM_003442
|
ZNF143
|
zinc finger protein 143
|
|
chr16_+_2802634
|
0.580
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr19_-_49540072
|
0.577
|
NM_033377
|
CGB1
CGB2
|
chorionic gonadotropin, beta polypeptide 1
chorionic gonadotropin, beta polypeptide 2
|
|
chrX_-_128977291
|
0.576
|
NM_001008222
|
ZDHHC9
|
zinc finger, DHHC-type containing 9
|
|
chr9_-_77643024
|
0.576
|
|
C9orf41
|
chromosome 9 open reading frame 41
|
|
chr6_+_148663953
|
0.574
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr16_+_19079220
|
0.573
|
NM_001190983
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr6_+_161412807
|
0.570
|
NM_005922
NM_006724
|
MAP3K4
|
mitogen-activated protein kinase kinase kinase 4
|
|
chr7_-_35734762
|
0.569
|
NM_022373
|
HERPUD2
|
HERPUD family member 2
|
|
chr7_+_129710401
|
0.568
|
|
KLHDC10
|
kelch domain containing 10
|
|
chrX_+_149737083
|
0.564
|
|
MTM1
|
myotubularin 1
|
|
chr1_+_179262848
|
0.564
|
NM_001252511
NM_001252512
NM_003101
|
SOAT1
|
sterol O-acyltransferase 1
|
|
chr19_+_57862633
|
0.559
|
NM_020657
|
ZNF304
|
zinc finger protein 304
|
|
chr6_+_35227478
|
0.559
|
NM_003427
|
ZNF76
|
zinc finger protein 76
|
|
chr9_+_111696720
|
0.557
|
|
FAM206A
|
family with sequence similarity 206, member A
|
|
chr16_-_46655262
|
0.557
|
|
SHCBP1
|
SHC SH2-domain binding protein 1
|
|
chr12_-_21654484
|
0.556
|
NM_002907
NM_032941
|
RECQL
|
RecQ protein-like (DNA helicase Q1-like)
|
|
chr1_+_40627055
|
0.556
|
|
RLF
|
rearranged L-myc fusion
|
|
chr10_+_14920781
|
0.555
|
NM_001193424
NM_001193426
NM_001193425
NM_001193427
NM_024670
|
SUV39H2
|
suppressor of variegation 3-9 homolog 2 (Drosophila)
|
|
chr16_+_2802652
|
0.555
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|