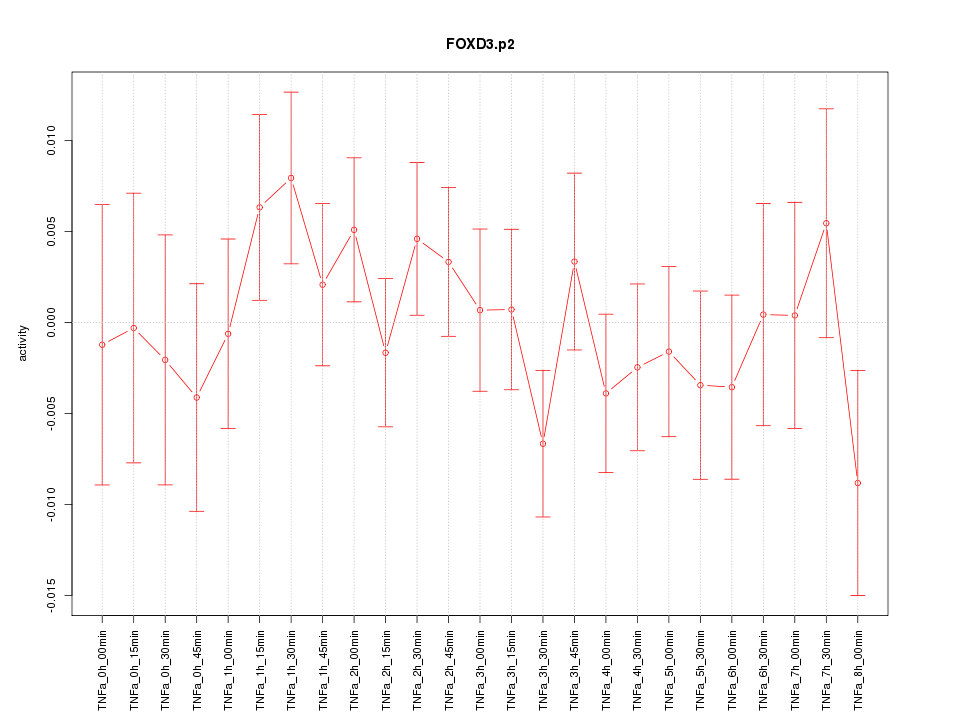
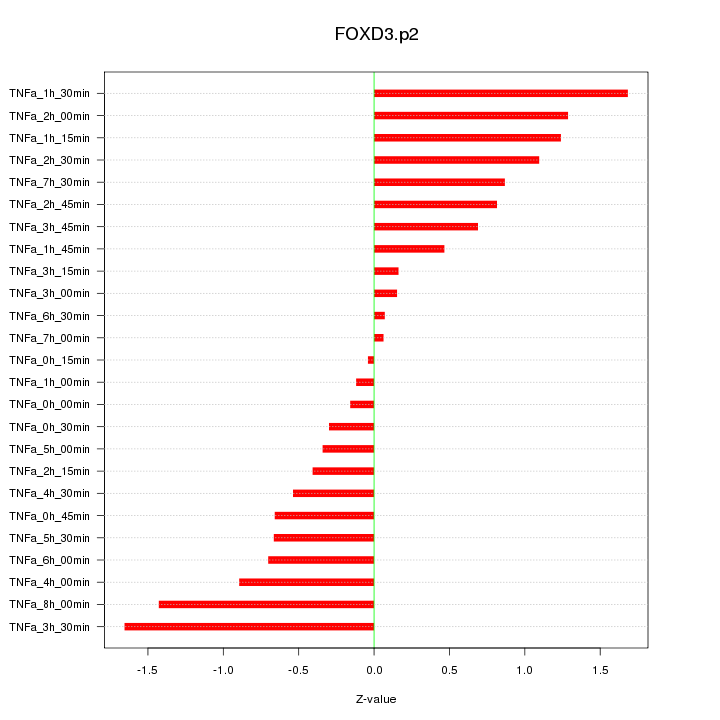
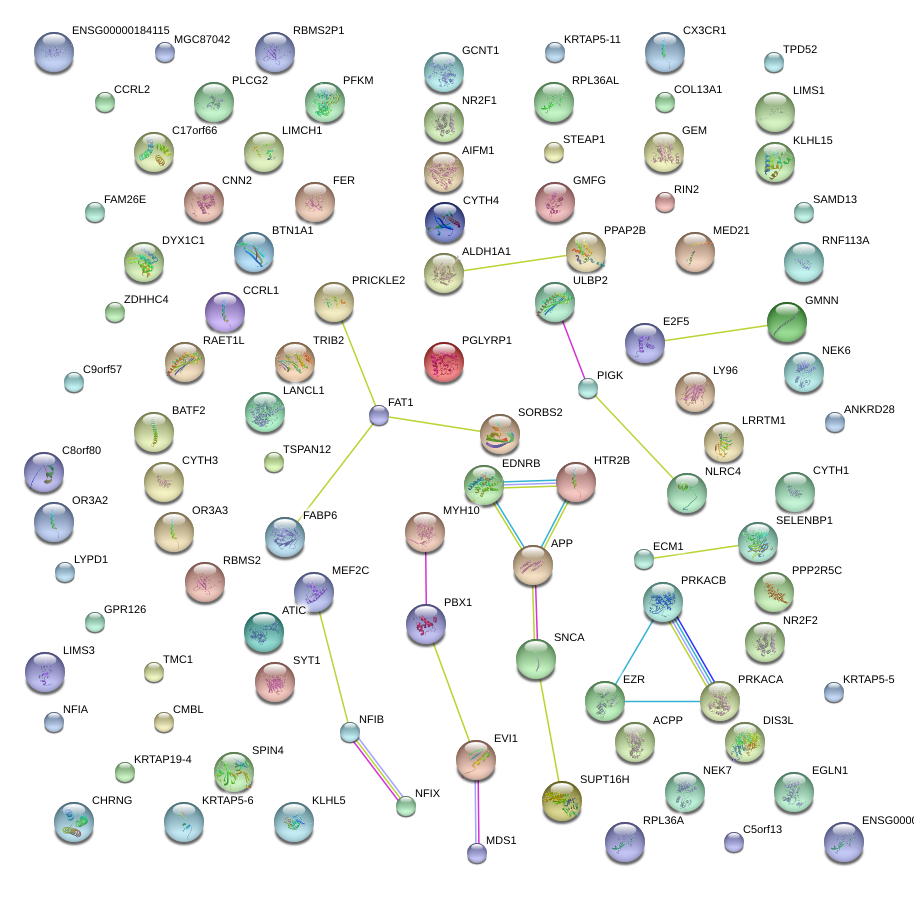
|
chr4_+_41614725
|
2.954
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr4_+_41614933
|
2.544
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr1_+_84630575
|
2.452
|
NM_001242860
NM_001242861
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr4_+_41614911
|
2.051
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr9_-_74675498
|
1.415
|
NM_001128618
|
C9orf57
|
chromosome 9 open reading frame 57
|
|
chr1_+_84764048
|
1.280
|
NM_001010971
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr1_-_57045235
|
1.275
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr3_-_64211111
|
1.228
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr21_-_27512668
|
1.216
|
NM_001136016
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr3_+_46448678
|
1.173
|
NM_003965
NM_001130910
|
CCRL2
|
chemokine (C-C motif) receptor-like 2
|
|
chr21_-_31869427
|
1.146
|
NM_181610
|
KRTAP19-4
|
keratin associated protein 19-4
|
|
chr4_-_186577858
|
1.139
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_+_84630374
|
1.129
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr1_+_84630014
|
1.128
|
NM_001242857
NM_001242858
NM_001242859
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr4_-_90759446
|
1.090
|
NM_001146055
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr5_-_111091947
|
1.057
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr2_+_109204777
|
1.048
|
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr1_-_57045214
|
1.023
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr4_-_186578122
|
0.935
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr5_-_10308167
|
0.899
|
NM_138809
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas)
|
|
chr5_+_159614373
|
0.875
|
NM_001130958
|
FABP6
|
fatty acid binding protein 6, ileal
|
|
chr2_-_133429069
|
0.868
|
NM_001077427
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr1_-_57045129
|
0.837
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr12_+_27175501
|
0.836
|
|
MED21
|
mediator complex subunit 21
|
|
chr6_+_26501448
|
0.833
|
NM_001732
|
BTN1A1
|
butyrophilin, subfamily 1, member A1
|
|
chr1_+_84609941
|
0.832
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr18_-_22930826
|
0.815
|
|
|
|
|
chr16_+_81812897
|
0.807
|
NM_002661
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr20_+_19870209
|
0.801
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr9_-_14086901
|
0.777
|
|
NFIB
|
nuclear factor I/B
|
|
chr5_-_88179278
|
0.745
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr5_-_997408
|
0.728
|
NM_001242737
|
LOC100506688
|
uncharacterized LOC100506688
|
|
chr2_+_12857885
|
0.724
|
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr9_+_79115548
|
0.718
|
NM_001097636
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr16_+_81812899
|
0.717
|
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr4_-_187644983
|
0.701
|
NM_005245
|
FAT1
|
FAT tumor suppressor homolog 1 (Drosophila)
|
|
chr2_+_216176808
|
0.692
|
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr8_+_74903563
|
0.692
|
NM_001195797
NM_015364
|
LY96
|
lymphocyte antigen 96
|
|
chr3_-_15839674
|
0.688
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr8_-_80993009
|
0.672
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr1_-_77685076
|
0.645
|
NM_005482
|
PIGK
|
phosphatidylinositol glycan anchor biosynthesis, class K
|
|
chr17_-_34195745
|
0.640
|
NM_152781
|
C17orf66
|
chromosome 17 open reading frame 66
|
|
chr17_-_8424217
|
0.636
|
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr2_-_32490688
|
0.636
|
NM_001199139
NM_021209
|
NLRC4
|
NLR family, CARD domain containing 4
|
|
chr9_-_75567915
|
0.632
|
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr1_-_57044980
|
0.625
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr2_-_80531486
|
0.624
|
NM_178839
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1
|
|
chr7_+_89783688
|
0.622
|
NM_012449
|
STEAP1
|
six transmembrane epithelial antigen of the prostate 1
|
|
chr12_+_48513012
|
0.610
|
NM_000289
|
PFKM
|
phosphofructokinase, muscle
|
|
chr15_-_55800431
|
0.609
|
NM_001033559
NM_001033560
NM_130810
|
DYX1C1
|
dyslexia susceptibility 1 candidate 1
|
|
chr1_+_164528586
|
0.603
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr3_+_132036210
|
0.602
|
NM_001099
NM_001134194
|
ACPP
|
acid phosphatase, prostate
|
|
chr1_+_84768333
|
0.601
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr14_+_102276125
|
0.597
|
NM_002719
NM_178586
NM_178587
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr6_+_142623364
|
0.596
|
|
GPR126
|
G protein-coupled receptor 126
|
|
chr7_-_120498128
|
0.591
|
NM_012338
|
TSPAN12
|
tetraspanin 12
|
|
chr2_+_216176818
|
0.591
|
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chrX_-_62570968
|
0.585
|
NM_001012968
|
SPIN4
|
spindlin family, member 4
|
|
chr2_+_216176678
|
0.581
|
NM_004044
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr12_+_79258517
|
0.578
|
|
SYT1
|
synaptotagmin I
|
|
chr8_-_95274540
|
0.578
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr9_+_127054246
|
0.572
|
NM_001166170
NM_001166171
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr20_+_19915716
|
0.567
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr19_-_39826676
|
0.566
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr9_-_75568232
|
0.563
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr12_+_56915587
|
0.562
|
NM_002898
|
RBMS2
|
RNA binding motif, single stranded interacting protein 2
|
|
chr12_+_79258448
|
0.560
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr2_+_109204745
|
0.551
|
NM_004987
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr17_-_3182267
|
0.549
|
NM_002551
|
OR3A2
|
olfactory receptor, family 3, subfamily A, member 2
|
|
chr2_-_231989755
|
0.547
|
NM_000867
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B
|
|
chr2_+_109204904
|
0.540
|
NM_001193488
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr6_+_116832807
|
0.535
|
NM_153711
|
FAM26E
|
family with sequence similarity 26, member E
|
|
chr1_-_151345156
|
0.524
|
NM_003944
|
SELENBP1
|
selenium binding protein 1
|
|
chr7_+_6617035
|
0.514
|
NM_001134387
NM_001134388
NM_001134389
NM_018106
|
ZDHHC4
|
zinc finger, DHHC-type containing 4
|
|
chr10_+_71561687
|
0.512
|
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr2_-_211341374
|
0.501
|
NM_001136574
NM_006055
NM_001136575
|
LANCL1
|
LanC lantibiotic synthetase component C-like 1 (bacterial)
|
|
chr8_-_27941387
|
0.492
|
NM_001010906
|
C8orf80
|
chromosome 8 open reading frame 80
|
|
chrX_-_24045302
|
0.485
|
NM_030624
|
KLHL15
|
kelch-like 15 (Drosophila)
|
|
chr15_+_96876568
|
0.481
|
NM_001145157
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr13_-_78493902
|
0.479
|
NM_001201397
|
EDNRB
|
endothelin receptor type B
|
|
chr7_+_89783789
|
0.468
|
|
STEAP1
|
six transmembrane epithelial antigen of the prostate 1
|
|
chr8_+_86099909
|
0.463
|
NM_001083589
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr1_+_150480486
|
0.456
|
NM_001202858
NM_004425
NM_022664
|
ECM1
|
extracellular matrix protein 1
|
|
chr22_+_37678565
|
0.450
|
|
CYTH4
|
cytohesin 4
|
|
chr11_-_16996559
|
0.449
|
|
RPL36A
|
ribosomal protein L36a
|
|
chr5_-_88199868
|
0.440
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr4_-_186877869
|
0.439
|
NM_021069
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_-_159240398
|
0.436
|
|
EZR
|
ezrin
|
|
chr22_+_37678423
|
0.435
|
NM_013385
|
CYTH4
|
cytohesin 4
|
|
chrX_-_129272014
|
0.432
|
NM_001130846
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chr11_+_1718424
|
0.431
|
NM_001012416
|
KRTAP5-6
|
keratin associated protein 5-6
|
|
chr1_+_61547388
|
0.426
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr6_+_150263120
|
0.423
|
NM_025217
|
ULBP2
|
UL16 binding protein 2
|
|
chr9_+_75136716
|
0.423
|
NM_138691
|
TMC1
|
transmembrane channel-like 1
|
|
chr15_+_66585632
|
0.423
|
NM_133375
|
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like
|
|
chr6_-_159240399
|
0.420
|
|
EZR
|
ezrin
|
|
chr3_+_132316080
|
0.420
|
NM_016557
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr10_+_71561643
|
0.416
|
NM_001130103
NM_080798
NM_080800
NM_080801
NM_080802
NM_080805
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr5_+_108083522
|
0.411
|
NM_005246
|
FER
|
fer (fps/fes related) tyrosine kinase
|
|
chr8_-_95274520
|
0.401
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr12_+_27175482
|
0.396
|
NM_004264
|
MED21
|
mediator complex subunit 21
|
|
chr19_+_1026271
|
0.394
|
NM_004368
NM_201277
|
CNN2
|
calponin 2
|
|
chr14_-_21852085
|
0.393
|
|
SUPT16H
|
suppressor of Ty 16 homolog (S. cerevisiae)
|
|
chr2_+_233404436
|
0.393
|
NM_005199
|
CHRNG
|
cholinergic receptor, nicotinic, gamma
|
|
chr1_-_231560789
|
0.393
|
NM_022051
|
EGLN1
|
egl nine homolog 1 (C. elegans)
|
|
chr3_-_168864066
|
0.391
|
NM_001105077
NM_001163999
NM_005241
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr19_-_49220213
|
0.389
|
NM_182574
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr19_-_39322411
|
0.388
|
NM_001398
|
ECH1
|
enoyl CoA hydratase 1, peroxisomal
|
|
chr15_-_66648991
|
0.385
|
NM_017858
|
TIPIN
|
TIMELESS interacting protein
|
|
chr11_+_87032428
|
0.383
|
|
TMEM135
|
transmembrane protein 135
|
|
chr2_-_96150478
|
0.381
|
NM_001164464
|
TRIM43B
|
tripartite motif containing 43B
|
|
chr1_+_74701070
|
0.376
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr11_+_86085777
|
0.372
|
NM_001156474
NM_021827
|
CCDC81
|
coiled-coil domain containing 81
|
|
chr11_-_88070840
|
0.371
|
|
CTSC
|
cathepsin C
|
|
chr3_-_18480203
|
0.368
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr2_-_183387063
|
0.363
|
NM_005019
NM_001003683
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr4_-_186732208
|
0.363
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_+_101185195
|
0.362
|
NM_001078
NM_001199834
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr12_-_124068864
|
0.359
|
|
|
|
|
chr8_-_17579729
|
0.359
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr2_-_158182154
|
0.358
|
NM_020711
|
ERMN
|
ermin, ERM-like protein
|
|
chr12_+_105153258
|
0.356
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr19_-_11689765
|
0.355
|
NM_001111035
NM_001111036
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr14_+_102276279
|
0.355
|
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr10_+_5238756
|
0.354
|
NM_001818
|
AKR1C4
|
aldo-keto reductase family 1, member C4 (chlordecone reductase; 3-alpha hydroxysteroid dehydrogenase, type I; dihydrodiol dehydrogenase 4)
|
|
chrX_-_2882288
|
0.350
|
NM_000047
|
ARSE
|
arylsulfatase E (chondrodysplasia punctata 1)
|
|
chr8_-_17658385
|
0.349
|
NM_001001924
NM_001001925
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr16_+_77756388
|
0.346
|
NM_001105663
NM_001243657
NM_001243660
NM_001243661
|
NUDT7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7
|
|
chr10_+_115310589
|
0.342
|
NM_001177660
|
HABP2
|
hyaluronan binding protein 2
|
|
chr4_-_102267952
|
0.342
|
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr11_-_88070901
|
0.339
|
|
CTSC
|
cathepsin C
|
|
chr12_-_14996381
|
0.339
|
NM_021071
|
ART4
|
ADP-ribosyltransferase 4 (Dombrock blood group)
|
|
chrX_-_63005212
|
0.337
|
NM_001173479
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr13_+_114239587
|
0.335
|
|
TFDP1
|
transcription factor Dp-1
|
|
chr14_-_61748193
|
0.334
|
|
|
|
|
chr2_-_119605758
|
0.332
|
NM_001426
|
EN1
|
engrailed homeobox 1
|
|
chr21_+_46293925
|
0.331
|
|
|
|
|
chr4_-_102268626
|
0.331
|
NM_000944
NM_001130691
NM_001130692
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr12_-_8088629
|
0.329
|
NM_006931
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3
|
|
chr12_+_20963637
|
0.329
|
NM_019844
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3
|
|
chr2_-_80531716
|
0.326
|
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1
|
|
chr2_+_237478379
|
0.323
|
NM_020311
|
CXCR7
|
chemokine (C-X-C motif) receptor 7
|
|
chr16_-_28437663
|
0.323
|
NM_001199142
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C
|
|
chr12_+_54892567
|
0.323
|
NM_001184976
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr15_+_42696957
|
0.319
|
NM_173089
NM_173090
|
CAPN3
|
calpain 3, (p94)
|
|
chr5_-_10307873
|
0.318
|
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas)
|
|
chr3_-_151987332
|
0.318
|
|
LOC401093
|
uncharacterized LOC401093
|
|
chr1_+_84767288
|
0.314
|
NM_001134663
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr5_+_139028483
|
0.313
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr15_+_58430394
|
0.313
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chr16_+_28699878
|
0.313
|
NM_001199142
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C
|
|
chr2_-_165424993
|
0.313
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr20_-_5107185
|
0.312
|
NM_002592
|
PCNA
|
proliferating cell nuclear antigen
|
|
chrX_+_55744049
|
0.312
|
NM_006064
NM_016656
|
RRAGB
|
Ras-related GTP binding B
|
|
chr16_+_67381252
|
0.310
|
NM_001161575
|
LRRC36
|
leucine rich repeat containing 36
|
|
chr10_-_116286570
|
0.309
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr13_+_97874704
|
0.308
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr1_-_114429996
|
0.306
|
|
BCL2L15
|
BCL2-like 15
|
|
chr13_+_103459495
|
0.305
|
NM_001204425
|
BIVM-ERCC5
BIVM
|
BIVM-ERCC5 readthrough
basic, immunoglobulin-like variable motif containing
|
|
chr1_+_164528914
|
0.305
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr13_+_99058147
|
0.302
|
|
|
|
|
chr2_-_219031646
|
0.299
|
NM_000634
|
CXCR1
|
chemokine (C-X-C motif) receptor 1
|
|
chr3_-_149051258
|
0.298
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr19_-_14785626
|
0.296
|
NM_032571
|
EMR3
|
egf-like module containing, mucin-like, hormone receptor-like 3
|
|
chr16_-_90076528
|
0.295
|
NM_024043
|
DBNDD1
|
dysbindin (dystrobrevin binding protein 1) domain containing 1
|
|
chr17_-_5138049
|
0.291
|
NM_207103
|
SCIMP
|
SLP adaptor and CSK interacting membrane protein
|
|
chr5_+_156696351
|
0.288
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr2_-_113012601
|
0.287
|
NM_032494
|
ZC3H8
|
zinc finger CCCH-type containing 8
|
|
chr18_+_77160243
|
0.287
|
NM_172387
NM_172389
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr3_+_11267666
|
0.286
|
NM_001098211
|
HRH1
|
histamine receptor H1
|
|
chr9_-_14398981
|
0.286
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr10_+_225932
|
0.285
|
NM_001202467
NM_001202468
|
ZMYND11
|
zinc finger, MYND-type containing 11
|
|
chr2_+_96257765
|
0.285
|
NM_138800
|
TRIM43
|
tripartite motif containing 43
|
|
chr11_-_17191287
|
0.284
|
NM_002645
|
PIK3C2A
|
phosphoinositide-3-kinase, class 2, alpha polypeptide
|
|
chr19_+_36726883
|
0.283
|
|
ZNF146
|
zinc finger protein 146
|
|
chr22_+_25423940
|
0.283
|
NM_001145206
|
KIAA1671
|
KIAA1671
|
|
chr5_+_159436143
|
0.282
|
|
TTC1
|
tetratricopeptide repeat domain 1
|
|
chr9_+_27109138
|
0.282
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr6_+_26104103
|
0.281
|
NM_003542
|
HIST2H4B
HIST1H4C
|
histone cluster 2, H4b
histone cluster 1, H4c
|
|
chr2_-_80531809
|
0.279
|
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1
|
|
chr17_+_67410825
|
0.278
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr6_+_148664124
|
0.278
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr20_-_45985411
|
0.278
|
NM_012408
NM_183047
NM_183048
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr15_+_49462408
|
0.276
|
NM_002044
|
GALK2
|
galactokinase 2
|
|
chr9_-_8733893
|
0.276
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr10_-_48390873
|
0.276
|
NM_002900
|
RBP3
|
retinol binding protein 3, interstitial
|
|
chr15_-_65321921
|
0.276
|
NM_139242
|
MTFMT
|
mitochondrial methionyl-tRNA formyltransferase
|
|
chr10_-_33623771
|
0.275
|
NM_001024628
NM_001024629
NM_001244972
NM_001244973
NM_003873
|
NRP1
|
neuropilin 1
|
|
chr1_+_36748164
|
0.274
|
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr1_-_169703178
|
0.273
|
NM_000450
|
SELE
|
selectin E
|
|
chr14_-_81405883
|
0.273
|
NM_152446
|
CEP128
|
centrosomal protein 128kDa
|
|
chr9_-_116163578
|
0.271
|
NM_000031
|
ALAD
|
aminolevulinate dehydratase
|
|
chr17_+_61086897
|
0.269
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr11_+_112047088
|
0.268
|
NM_001037290
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr9_-_14314036
|
0.268
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr11_-_107582733
|
0.267
|
NM_003063
|
SLN
|
sarcolipin
|
|
chr1_+_109102970
|
0.266
|
NM_001010883
|
FAM102B
|
family with sequence similarity 102, member B
|
|
chr1_-_92371558
|
0.265
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr16_+_11439279
|
0.262
|
NM_152308
|
RMI2
|
RMI2, RecQ mediated genome instability 2, homolog (S. cerevisiae)
|
|
chr16_+_4382224
|
0.262
|
NM_032575
|
GLIS2
|
GLIS family zinc finger 2
|