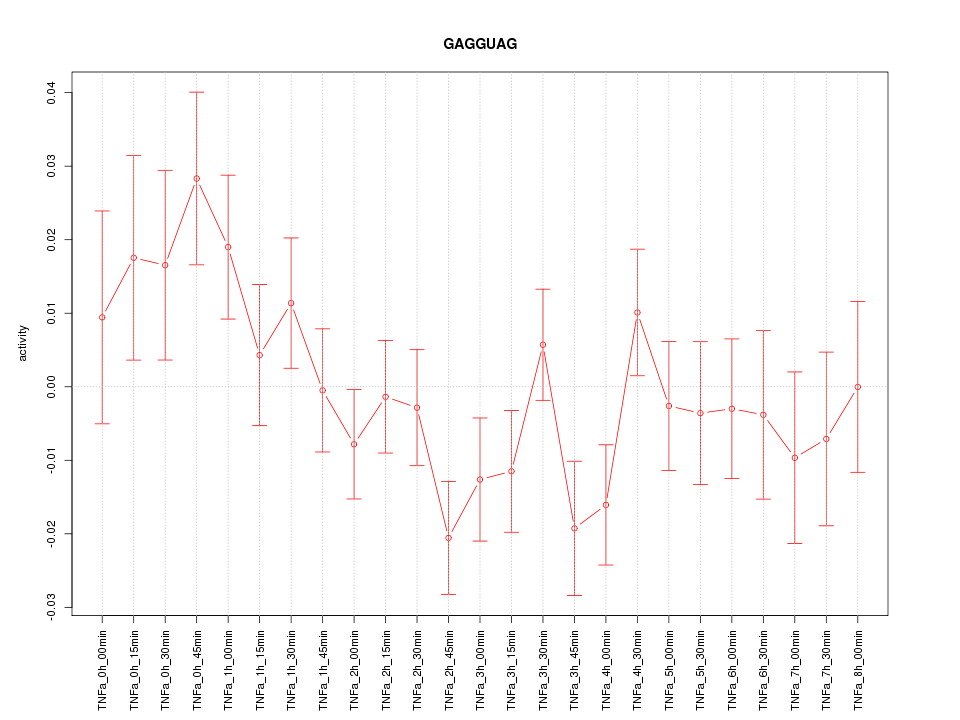
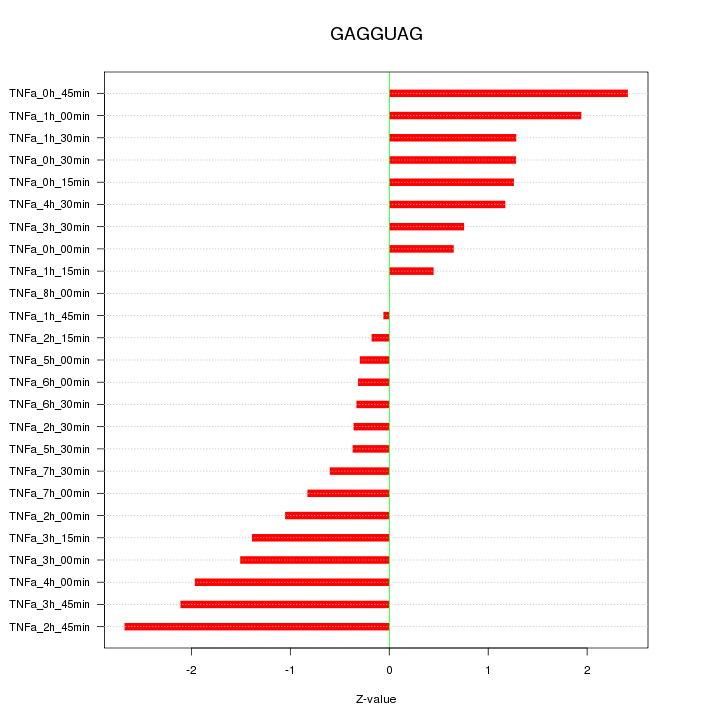
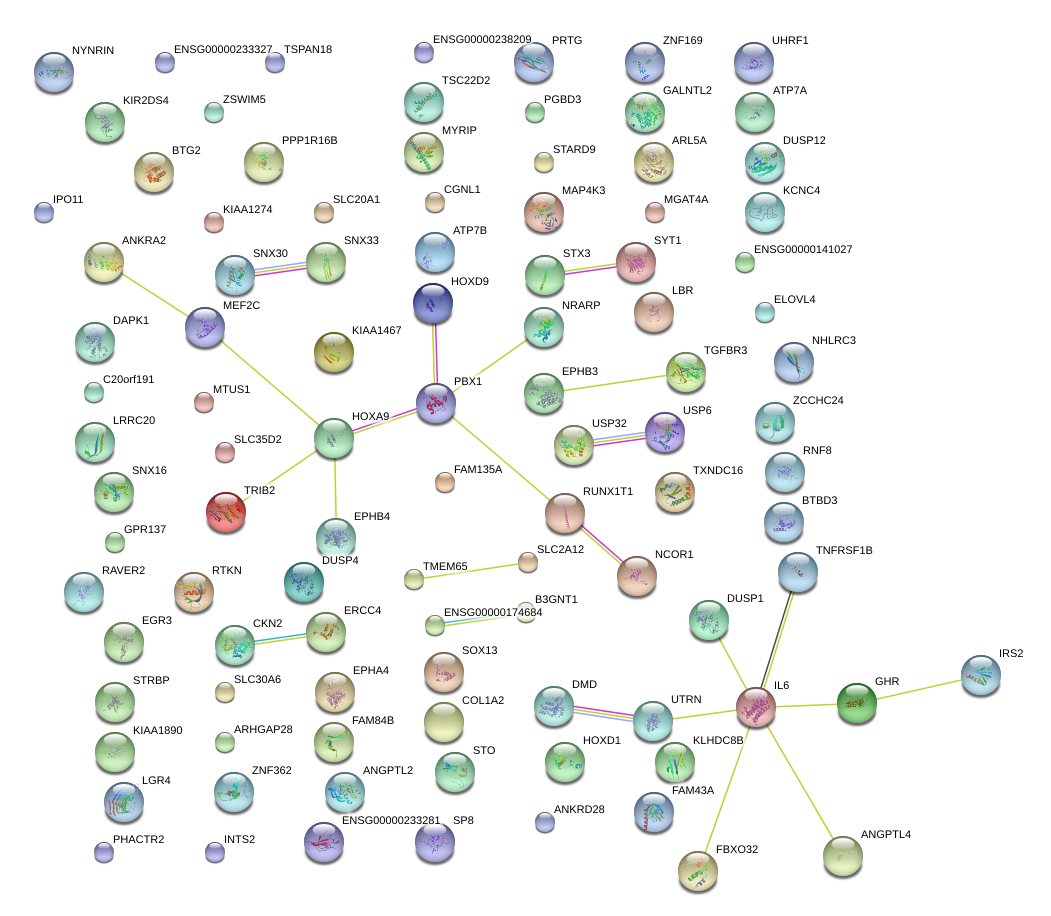
|
chr8_-_22549901
|
3.212
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr8_-_22550708
|
3.131
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr8_-_22550057
|
3.129
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr8_-_93029864
|
2.900
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93107881
|
2.818
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93111915
|
2.783
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_-_92371558
|
2.704
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr8_-_93107442
|
2.647
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_-_92351613
|
2.624
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr8_-_93115453
|
2.602
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr14_+_24867926
|
2.534
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr8_-_93075190
|
2.247
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr5_-_88179278
|
2.200
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr5_-_172198160
|
2.073
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr7_+_94023872
|
2.027
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr15_+_57668702
|
2.027
|
NM_001252335
NM_032866
|
CGNL1
|
cingulin-like 1
|
|
chr5_-_88199868
|
1.989
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr7_-_20826503
|
1.951
|
NM_182700
NM_198956
|
SP8
|
Sp8 transcription factor
|
|
chr2_+_12856813
|
1.950
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr8_-_125384916
|
1.874
|
NM_194291
|
TMEM65
|
transmembrane protein 65
|
|
chr2_-_152684963
|
1.870
|
NM_001037174
NM_012097
NM_177985
|
ARL5A
|
ADP-ribosylation factor-like 5A
|
|
chr1_-_45672225
|
1.869
|
NM_020883
|
ZSWIM5
|
zinc finger, SWIM-type containing 5
|
|
chr3_+_39851028
|
1.851
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr11_+_59522531
|
1.817
|
NM_001178040
NM_004177
|
STX3
|
syntaxin 3
|
|
chr8_-_124544376
|
1.698
|
NM_148177
|
FBXO32
|
F-box protein 32
|
|
chr8_-_124553448
|
1.690
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr2_-_39664129
|
1.663
|
NM_003618
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3
|
|
chr3_-_15900928
|
1.643
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr3_+_16216155
|
1.640
|
NM_054110
|
GALNTL2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 2
|
|
chr20_+_37434294
|
1.627
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B
|
|
chr2_-_222436996
|
1.614
|
NM_004438
|
EPHA4
|
EPH receptor A4
|
|
chr10_-_50747080
|
1.582
|
NM_000124
|
ERCC6
PGBD3
|
excision repair cross-complementing rodent repair deficiency, complementation group 6
piggyBac transposable element derived 3
|
|
chr9_+_115513050
|
1.544
|
NM_001012994
|
SNX30
|
sorting nexin family member 30
|
|
chr1_-_225616556
|
1.540
|
NM_194442
|
LBR
|
lamin B receptor
|
|
chr2_-_74667553
|
1.538
|
NM_001015056
NM_033046
|
RTKN
|
rhotekin
|
|
chr5_-_88119604
|
1.534
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr13_-_52585472
|
1.512
|
NM_000053
NM_001005918
NM_001243182
|
ATP7B
|
ATPase, Cu++ transporting, beta polypeptide
|
|
chr7_+_22766765
|
1.510
|
NM_000600
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr18_+_6834431
|
1.462
|
NM_001010000
|
ARHGAP28
|
Rho GTPase activating protein 28
|
|
chr9_+_90112754
|
1.439
|
NM_004938
|
DAPK1
|
death-associated protein kinase 1
|
|
chr7_-_27205148
|
1.402
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr12_+_13197284
|
1.402
|
NM_020853
|
KIAA1467
|
KIAA1467
|
|
chr9_-_140196702
|
1.391
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr8_-_127570710
|
1.387
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr9_+_123850573
|
1.379
|
NM_007018
|
CNTRL
|
centriolin
|
|
chr3_-_15839674
|
1.377
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr15_+_42867708
|
1.372
|
NM_020759
|
STARD9
|
StAR-related lipid transfer (START) domain containing 9
|
|
chr13_+_39612440
|
1.333
|
NM_001012754
NM_001017370
|
NHLRC3
|
NHL repeat containing 3
|
|
chr1_+_12227050
|
1.319
|
NM_001066
|
TNFRSF1B
|
tumor necrosis factor receptor superfamily, member 1B
|
|
chr2_-_74669055
|
1.315
|
NM_001015055
|
RTKN
|
rhotekin
|
|
chr2_-_99347588
|
1.314
|
NM_012214
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr1_+_203274646
|
1.241
|
NM_006763
|
BTG2
|
BTG family, member 2
|
|
chr1_+_164528586
|
1.236
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr10_+_72238529
|
1.215
|
NM_014431
|
KIAA1274
|
KIAA1274
|
|
chr15_-_56035176
|
1.193
|
NM_173814
|
PRTG
|
protogenin
|
|
chr1_-_225615787
|
1.176
|
NM_002296
|
LBR
|
lamin B receptor
|
|
chr6_-_80656870
|
1.158
|
NM_022726
|
ELOVL4
|
ELOVL fatty acid elongase 4
|
|
chr8_-_82754427
|
1.158
|
NM_022133
NM_152836
NM_152837
|
SNX16
|
sorting nexin 16
|
|
chr16_+_14013989
|
1.150
|
NM_005236
|
ERCC4
|
excision repair cross-complementing rodent repair deficiency, complementation group 4
|
|
chr2_+_113403433
|
1.147
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr3_+_150126705
|
1.138
|
NM_014779
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chr5_-_72861376
|
1.133
|
NM_023039
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2
|
|
chr6_+_143999058
|
1.133
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr1_+_33722158
|
1.096
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chrX_-_33229428
|
1.093
|
NM_004006
|
DMD
|
dystrophin
|
|
chr6_+_37321719
|
1.089
|
NM_003958
NM_183078
|
RNF8
|
ring finger protein 8
|
|
chr3_-_56717132
|
1.087
|
NM_001112736
|
FAM208A
|
family with sequence similarity 208, member A
|
|
chr5_+_61714710
|
1.076
|
NM_001134779
|
IPO11
|
importin 11
|
|
chr3_+_49208979
|
1.066
|
NM_173546
|
KLHDC8B
|
kelch domain containing 8B
|
|
chr9_-_129884998
|
1.063
|
NM_012098
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr5_+_176560056
|
1.054
|
NM_172349
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr11_-_66115016
|
1.049
|
NM_006876
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr1_+_204042191
|
1.048
|
NM_005686
|
SOX13
|
SRY (sex determining region Y)-box 13
|
|
chr6_-_134373618
|
1.046
|
NM_145176
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12
|
|
chr6_+_143929316
|
1.031
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr8_-_29206321
|
1.027
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr3_-_15798057
|
1.022
|
NM_001195099
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr13_-_110438896
|
1.019
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr2_+_177053306
|
1.019
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr5_+_61708565
|
1.015
|
NM_016338
|
IPO11
|
importin 11
|
|
chr12_+_79257772
|
1.010
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr12_+_79439432
|
1.007
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr2_+_32390902
|
0.998
|
NM_001193513
NM_001193514
NM_001193515
NM_017964
|
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6
|
|
chr5_+_42549657
|
0.985
|
NM_001242404
|
GHR
|
growth hormone receptor
|
|
chr5_+_42565963
|
0.979
|
NM_001242460
NM_001242461
NM_001242462
|
GHR
|
growth hormone receptor
|
|
chr20_+_11871364
|
0.977
|
NM_181443
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr1_+_110753313
|
0.967
|
NM_001039574
NM_004978
|
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4
|
|
chr10_-_72141347
|
0.966
|
NM_207119
|
LRRC20
|
leucine rich repeat containing 20
|
|
chr17_-_60005309
|
0.966
|
NM_020748
|
INTS2
|
integrator complex subunit 2
|
|
chr3_+_194406614
|
0.963
|
NM_153690
|
FAM43A
|
family with sequence similarity 43, member A
|
|
chr9_-_126030854
|
0.962
|
NM_001171137
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr9_-_99145991
|
0.960
|
NM_007001
|
SLC35D2
|
solute carrier family 35, member D2
|
|
chr5_+_42425093
|
0.956
|
NM_001242401
|
GHR
|
growth hormone receptor
|
|
chr5_+_42550181
|
0.947
|
NM_001242405
|
GHR
|
growth hormone receptor
|
|
chr17_+_5031686
|
0.946
|
NM_004505
|
USP6
|
ubiquitin specific peptidase 6 (Tre-2 oncogene)
|
|
chr1_+_65210724
|
0.946
|
NM_018211
|
RAVER2
|
ribonucleoprotein, PTB-binding 2
|
|
chr19_+_4910339
|
0.943
|
NM_013282
|
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chr11_+_44785975
|
0.926
|
NM_130783
|
TSPAN18
|
tetraspanin 18
|
|
chr10_-_81205091
|
0.926
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chrX_-_32173585
|
0.925
|
NM_004013
NM_004020
NM_004021
NM_004022
NM_004023
|
DMD
|
dystrophin
|
|
chr3_-_56698073
|
0.919
|
NM_015224
|
FAM208A
|
family with sequence similarity 208, member A
|
|
chr17_-_16118789
|
0.919
|
NM_006311
|
NCOR1
|
nuclear receptor corepressor 1
|
|
chr9_-_126030792
|
0.918
|
NM_018387
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr8_-_17555158
|
0.917
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr6_+_71123105
|
0.915
|
NM_001105531
NM_020819
|
FAM135A
|
family with sequence similarity 135, member A
|
|
chr11_-_27494232
|
0.914
|
NM_018490
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4
|
|
chr16_-_4166185
|
0.912
|
NM_001116
|
ADCY9
|
adenylate cyclase 9
|
|
chr12_+_79258448
|
0.910
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr5_+_42424553
|
0.906
|
NM_001242399
NM_001242400
|
GHR
|
growth hormone receptor
|
|
chr4_-_102268626
|
0.900
|
NM_000944
NM_001130691
NM_001130692
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr9_-_10612722
|
0.900
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr10_-_72142344
|
0.899
|
NM_018205
NM_018239
|
LRRC20
|
leucine rich repeat containing 20
|
|
chr21_-_28217692
|
0.893
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr16_+_53088791
|
0.887
|
NM_025134
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr9_-_8733893
|
0.885
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr11_+_86748870
|
0.881
|
NM_001168724
NM_022918
|
TMEM135
|
transmembrane protein 135
|
|
chr14_+_24899139
|
0.881
|
NM_015299
|
KHNYN
|
KH and NYN domain containing
|
|
chr5_+_42548144
|
0.881
|
NM_001242402
|
GHR
|
growth hormone receptor
|
|
chrX_-_33146543
|
0.867
|
NM_004010
NM_004009
|
DMD
|
dystrophin
|
|
chr8_-_17658385
|
0.864
|
NM_001001924
NM_001001925
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr5_+_42548314
|
0.859
|
NM_001242403
|
GHR
|
growth hormone receptor
|
|
chr9_+_128510477
|
0.856
|
NM_001134778
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr6_-_8064642
|
0.853
|
NM_001199322
NM_001199323
NM_201280
|
MUTED
|
muted homolog (mouse)
|
|
chr3_+_183873253
|
0.852
|
NM_004423
|
DVL3
|
dishevelled, dsh homolog 3 (Drosophila)
|
|
chr16_+_18995255
|
0.851
|
NM_024847
|
TMC7
|
transmembrane channel-like 7
|
|
chr3_-_69062744
|
0.851
|
NM_173654
|
C3orf64
|
chromosome 3 open reading frame 64
|
|
chr8_-_17580024
|
0.851
|
NM_001166393
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr8_-_17579729
|
0.842
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr5_-_55008125
|
0.836
|
NM_173514
|
SLC38A9
|
solute carrier family 38, member 9
|
|
chr8_+_74206772
|
0.829
|
NM_172037
|
RDH10
|
retinol dehydrogenase 10 (all-trans)
|
|
chr11_+_134094545
|
0.828
|
NM_052875
|
VPS26B
|
vacuolar protein sorting 26 homolog B (S. pombe)
|
|
chr3_-_132441302
|
0.826
|
NM_153240
|
NPHP3-ACAD11
NPHP3
|
NPHP3-ACAD11 readthrough
nephronophthisis 3 (adolescent)
|
|
chrX_-_31526414
|
0.822
|
NM_004014
|
DMD
|
dystrophin
|
|
chrX_-_32430370
|
0.818
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr6_+_71136141
|
0.817
|
NM_001162529
|
FAM135A
|
family with sequence similarity 135, member A
|
|
chr8_-_29207795
|
0.817
|
NM_001394
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr8_-_12973752
|
0.816
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr1_+_161129229
|
0.805
|
NM_001014443
NM_012475
|
USP21
|
ubiquitin specific peptidase 21
|
|
chr1_-_204121144
|
0.802
|
NM_018208
|
ETNK2
|
ethanolamine kinase 2
|
|
chr10_-_102279590
|
0.798
|
NM_015490
|
SEC31B
|
SEC31 homolog B (S. cerevisiae)
|
|
chr13_-_96296919
|
0.797
|
NM_014934
NM_198968
|
DZIP1
|
DAZ interacting protein 1
|
|
chr16_+_18995508
|
0.796
|
NM_001160364
|
TMC7
|
transmembrane channel-like 7
|
|
chr2_-_70475563
|
0.796
|
NM_022037
NM_022173
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein
|
|
chr9_+_132597651
|
0.795
|
NM_001110303
NM_006676
|
USP20
|
ubiquitin specific peptidase 20
|
|
chr14_-_89883306
|
0.793
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr1_-_179112178
|
0.792
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr13_-_99174251
|
0.789
|
NM_003576
|
STK24
|
serine/threonine kinase 24
|
|
chr12_+_111843743
|
0.786
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr9_+_132597728
|
0.781
|
NM_001008563
|
USP20
|
ubiquitin specific peptidase 20
|
|
chr13_-_52733669
|
0.780
|
NM_152720
|
NEK3
|
NIMA (never in mitosis gene a)-related kinase 3
|
|
chr5_+_42423811
|
0.779
|
NM_000163
|
GHR
|
growth hormone receptor
|
|
chr13_-_33780142
|
0.778
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr13_-_33760215
|
0.776
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr4_+_106067754
|
0.775
|
NM_001127208
|
TET2
|
tet methylcytosine dioxygenase 2
|
|
chr20_-_62601140
|
0.769
|
NM_020713
|
ZNF512B
|
zinc finger protein 512B
|
|
chr12_-_59313304
|
0.769
|
NM_001136051
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr5_+_41904459
|
0.767
|
NM_175921
|
C5orf51
|
chromosome 5 open reading frame 51
|
|
chr13_-_52733995
|
0.766
|
NM_001146099
NM_002498
|
NEK3
|
NIMA (never in mitosis gene a)-related kinase 3
|
|
chr12_-_59314069
|
0.760
|
NM_153377
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr5_+_42565562
|
0.754
|
NM_001242406
|
GHR
|
growth hormone receptor
|
|
chr14_+_57857261
|
0.752
|
NM_001011713
|
NAA30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit
|
|
chr7_+_74072007
|
0.751
|
NM_001163636
NM_001518
NM_032999
NM_033000
NM_033001
|
GTF2I
|
general transcription factor IIi
|
|
chr9_+_128509616
|
0.749
|
NM_006195
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr17_+_61699786
|
0.741
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr20_+_11898536
|
0.740
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr5_+_150157507
|
0.727
|
NM_032947
|
C5orf62
|
chromosome 5 open reading frame 62
|
|
chr14_+_100150596
|
0.723
|
NM_006668
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1
|
|
chr13_-_99229226
|
0.722
|
NM_001032296
|
STK24
|
serine/threonine kinase 24
|
|
chr4_-_18023358
|
0.719
|
NM_001166139
NM_153686
|
LCORL
|
ligand dependent nuclear receptor corepressor-like
|
|
chr12_+_56915587
|
0.713
|
NM_002898
|
RBMS2
|
RNA binding motif, single stranded interacting protein 2
|
|
chr1_-_6296034
|
0.706
|
NM_012405
|
ICMT
|
isoprenylcysteine carboxyl methyltransferase
|
|
chr2_+_148602569
|
0.703
|
NM_001616
|
ACVR2A
|
activin A receptor, type IIA
|
|
chr17_-_58469577
|
0.702
|
NM_032582
|
USP32
|
ubiquitin specific peptidase 32
|
|
chr21_+_27107322
|
0.695
|
NM_002040
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa
|
|
chr3_+_107241782
|
0.694
|
NM_001142568
NM_020235
|
BBX
|
bobby sox homolog (Drosophila)
|
|
chr8_-_12990731
|
0.690
|
NM_006094
|
DLC1
|
deleted in liver cancer 1
|
|
chr16_+_66914380
|
0.687
|
NM_020786
|
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2
|
|
chrX_-_33357725
|
0.685
|
NM_000109
|
DMD
|
dystrophin
|
|
chr19_+_4909447
|
0.681
|
NM_001048201
|
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chr3_-_185542807
|
0.681
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr21_+_27107257
|
0.676
|
NM_001197297
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa
|
|
chr10_+_102106771
|
0.675
|
NM_005063
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr9_-_72287097
|
0.671
|
NM_001163
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1
|
|
chr12_-_42538432
|
0.666
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr12_-_48745020
|
0.663
|
NM_001172682
|
ZNF641
|
zinc finger protein 641
|
|
chrX_-_31284946
|
0.661
|
NM_004015
NM_004016
NM_004017
NM_004018
NM_004019
|
DMD
|
dystrophin
|
|
chr15_-_49338496
|
0.660
|
NM_001193489
NM_014701
|
SECISBP2L
|
SECIS binding protein 2-like
|
|
chr12_+_15475190
|
0.659
|
NM_002848
NM_030667
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr4_-_128887092
|
0.658
|
NM_152778
|
MFSD8
|
major facilitator superfamily domain containing 8
|
|
chr17_-_65241305
|
0.658
|
NM_014877
|
HELZ
|
helicase with zinc finger
|
|
chr9_+_136325086
|
0.648
|
NM_001135775
NM_001242369
NM_001242370
NM_017586
|
C9orf7
|
chromosome 9 open reading frame 7
|
|
chr12_+_15699277
|
0.642
|
NM_030668
NM_030669
NM_030670
NM_030671
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr6_+_147525493
|
0.639
|
NM_001127715
NM_139244
|
STXBP5
|
syntaxin binding protein 5 (tomosyn)
|
|
chr17_-_58603492
|
0.635
|
NM_006380
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2
|
|
chr2_+_109335906
|
0.633
|
NM_006267
|
RANBP2
|
RAN binding protein 2
|
|
chr1_+_154377668
|
0.633
|
NM_000565
NM_001206866
NM_181359
|
IL6R
|
interleukin 6 receptor
|
|
chr7_-_6312137
|
0.632
|
NM_004227
|
CYTH3
|
cytohesin 3
|
|
chr1_-_165325951
|
0.629
|
NM_177398
|
LMX1A
|
LIM homeobox transcription factor 1, alpha
|
|
chr20_+_278200
|
0.627
|
NM_033089
|
ZCCHC3
|
zinc finger, CCHC domain containing 3
|
|
chr13_-_33859835
|
0.623
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|