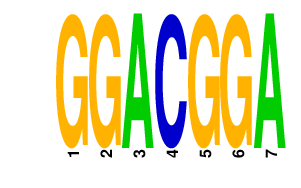
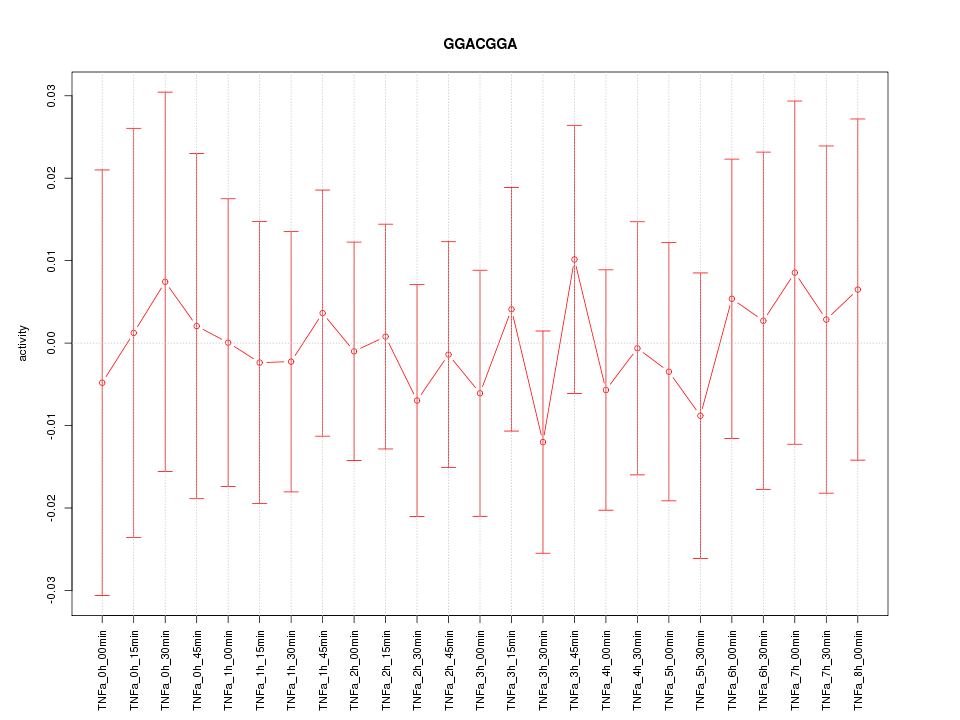
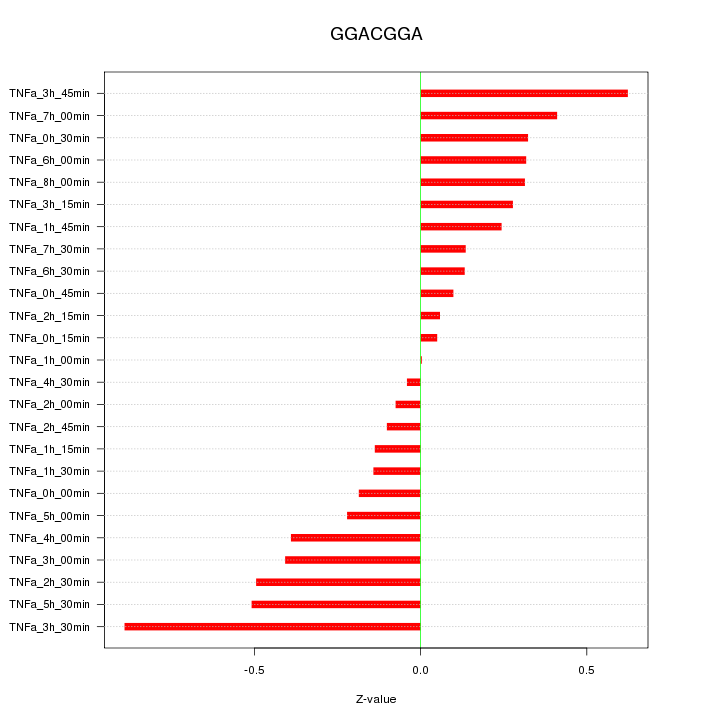
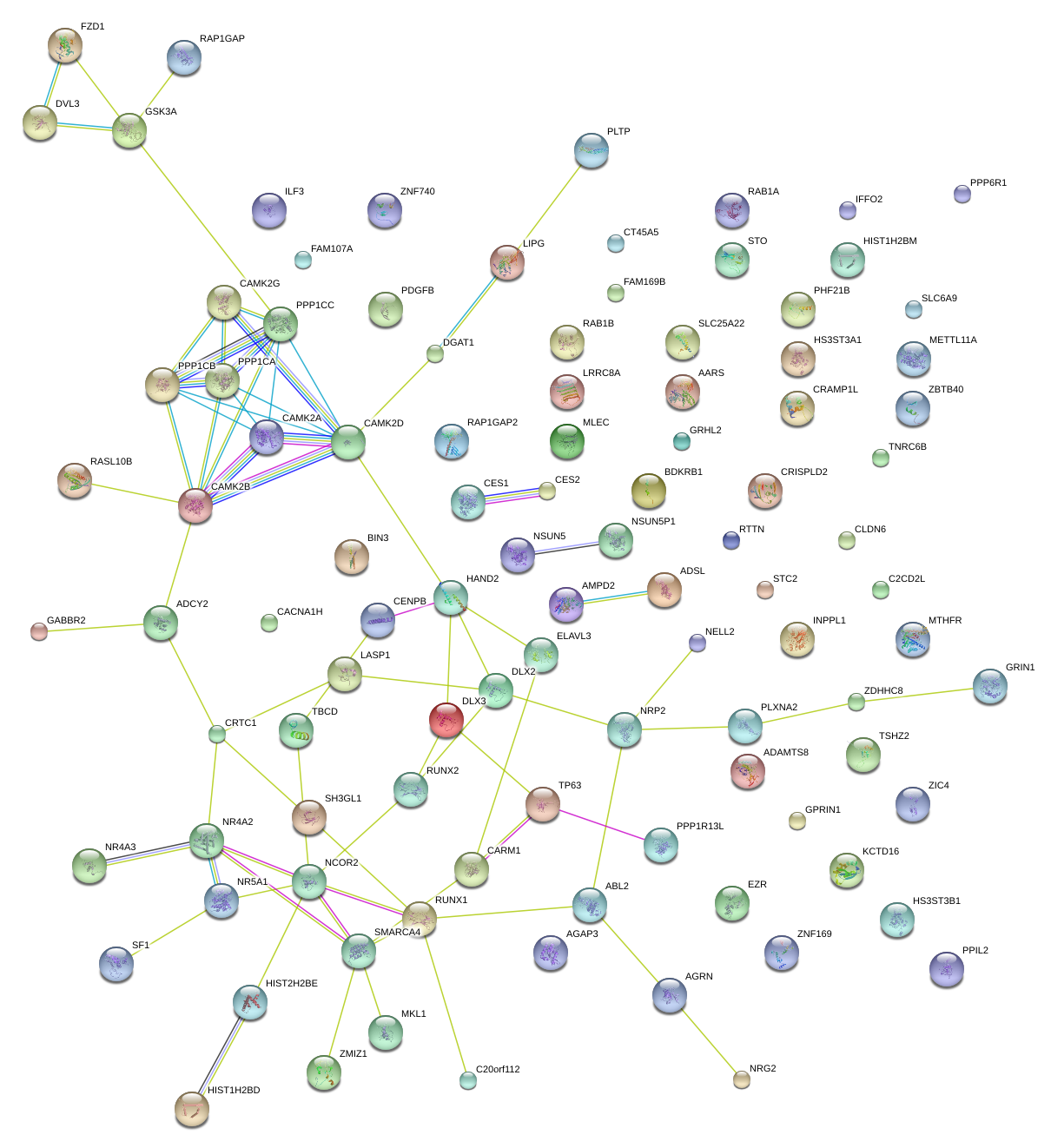
|
chr5_-_172756505
|
0.307
|
NM_003714
|
STC2
|
stanniocalcin 2
|
|
chr4_-_174451356
|
0.210
|
NM_021973
|
HAND2
|
heart and neural crest derivatives expressed 2
|
|
chr11_-_798196
|
0.175
|
NM_001191061
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22
|
|
chr11_-_797955
|
0.174
|
NM_001191060
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22
|
|
chr20_-_44541002
|
0.169
|
NM_001242920
NM_006227
NM_182676
|
PLTP
|
phospholipid transfer protein
|
|
chr1_-_44482996
|
0.152
|
NM_006934
NM_201649
|
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9
|
|
chr20_-_44539597
|
0.151
|
NM_001242921
|
PLTP
|
phospholipid transfer protein
|
|
chr19_-_45909566
|
0.139
|
NM_001142502
|
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like
|
|
chr11_-_796175
|
0.136
|
NM_024698
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22
|
|
chr19_-_45908288
|
0.132
|
NM_006663
|
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like
|
|
chr20_-_31071187
|
0.128
|
NM_080616
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr3_-_147123306
|
0.124
|
NM_001168379
|
ZIC4
|
Zic family member 4
|
|
chr3_-_147122070
|
0.123
|
NM_001168378
|
ZIC4
|
Zic family member 4
|
|
chr21_-_36421586
|
0.118
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr3_-_147124595
|
0.107
|
NM_001243256
NM_032153
|
ZIC4
|
Zic family member 4
|
|
chr5_+_143550395
|
0.102
|
NM_020768
|
KCTD16
|
potassium channel tetramerisation domain containing 16
|
|
chr16_+_1203240
|
0.102
|
NM_001005407
NM_021098
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr1_-_44497132
|
0.100
|
NM_001024845
|
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9
|
|
chr7_+_90893735
|
0.098
|
NM_003505
|
FZD1
|
frizzled family receptor 1
|
|
chr12_-_125052009
|
0.096
|
NM_001077261
NM_001206654
NM_006312
|
NCOR2
|
nuclear receptor corepressor 2
|
|
chr1_+_955468
|
0.091
|
NM_198576
|
AGRN
|
agrin
|
|
chr5_+_176560056
|
0.089
|
NM_172349
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr2_-_157189040
|
0.086
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr22_+_40440664
|
0.084
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr9_+_131644368
|
0.083
|
NM_001127245
NM_019594
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr22_-_45405429
|
0.078
|
NM_138415
|
PHF21B
|
PHD finger protein 21B
|
|
chr18_+_47088400
|
0.078
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr3_-_58563490
|
0.078
|
NM_001076778
NM_007177
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr19_+_10982244
|
0.077
|
NM_199141
|
CARM1
|
coactivator-associated arginine methyltransferase 1
|
|
chr22_-_45404785
|
0.075
|
NM_001242450
|
PHF21B
|
PHD finger protein 21B
|
|
chr22_-_32146119
|
0.074
|
NM_173566
|
PRR14L
|
proline rich 14-like
|
|
chr9_+_131644780
|
0.073
|
NM_001127244
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr22_-_45405780
|
0.073
|
NM_001135862
|
PHF21B
|
PHD finger protein 21B
|
|
chr12_+_121124922
|
0.070
|
NM_014730
|
MLEC
|
malectin
|
|
chr17_+_37026111
|
0.067
|
NM_006148
|
LASP1
|
LIM and SH3 protein 1
|
|
chr22_+_40573879
|
0.065
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr5_+_7396030
|
0.064
|
NM_020546
|
ADCY2
|
adenylate cyclase 2 (brain)
|
|
chr15_-_99057610
|
0.062
|
NM_182562
|
FAM169B
|
family with sequence similarity 169, member B
|
|
chr1_-_11866079
|
0.058
|
NM_005957
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr19_+_11071583
|
0.054
|
NM_001128844
NM_003072
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4
|
|
chr19_+_11071792
|
0.054
|
NM_001128849
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4
|
|
chr19_-_4400470
|
0.053
|
NM_001199943
NM_003025
|
SH3GL1
|
SH3-domain GRB2-like 1
|
|
chr17_-_48072566
|
0.053
|
NM_005220
|
DLX3
|
distal-less homeobox 3
|
|
chr22_-_39636906
|
0.050
|
NM_033016
|
PDGFB
|
platelet-derived growth factor beta polypeptide
|
|
chr11_-_64545933
|
0.050
|
NM_001178030
|
SF1
|
splicing factor 1
|
|
chr11_-_64545231
|
0.050
|
NM_001178031
|
SF1
|
splicing factor 1
|
|
chr11_+_71935824
|
0.048
|
NM_001567
|
INPPL1
|
inositol polyphosphate phosphatase-like 1
|
|
chr17_+_14204326
|
0.042
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr19_-_55769920
|
0.041
|
NM_014931
|
PPP6R1
|
protein phosphatase 6, regulatory subunit 1
|
|
chr12_+_53574506
|
0.041
|
NM_001004304
|
ZNF740
|
zinc finger protein 740
|
|
chr1_-_19282773
|
0.041
|
NM_001136265
|
IFFO2
|
intermediate filament family orphan 2
|
|
chr16_-_70323407
|
0.039
|
NM_001605
|
AARS
|
alanyl-tRNA synthetase
|
|
chr21_-_36260972
|
0.038
|
NM_001001890
NM_001122607
|
RUNX1
|
runt-related transcription factor 1
|
|
chr1_+_110162427
|
0.038
|
NM_139156
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr1_+_110163275
|
0.037
|
NM_004037
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr1_-_208417635
|
0.036
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr19_-_42746723
|
0.036
|
NM_019884
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr1_+_110168048
|
0.036
|
NM_203404
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr16_+_66968319
|
0.034
|
NM_003869
NM_198061
|
CES2
|
carboxylesterase 2
|
|
chr6_+_27782793
|
0.034
|
NM_003521
|
HIST1H2BM
|
histone cluster 1, H2bm
|
|
chr7_-_72722786
|
0.033
|
NM_001168347
NM_001168348
NM_018044
NM_148956
|
NSUN5
|
NOP2/Sun domain family, member 5
|
|
chr8_-_22526581
|
0.033
|
NM_018688
|
BIN3
|
bridging integrator 3
|
|
chr19_-_4400357
|
0.031
|
NM_001199944
|
SH3GL1
|
SH3-domain GRB2-like 1
|
|
chr7_-_44365019
|
0.030
|
NM_001220
NM_172078
NM_172079
NM_172080
NM_172081
NM_172082
NM_172083
NM_172084
|
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta
|
|
chr22_+_20119354
|
0.029
|
NM_001185024
NM_013373
|
ZDHHC8
|
zinc finger, DHHC-type containing 8
|
|
chr19_-_11591439
|
0.028
|
NM_001420
NM_032281
|
ELAVL3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr19_+_11094799
|
0.027
|
NM_001128845
NM_001128846
NM_001128847
NM_001128848
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4
|
|
chr20_-_3766905
|
0.027
|
NM_001810
|
CENPB
|
centromere protein B, 80kDa
|
|
chr22_-_39640956
|
0.027
|
NM_002608
|
PDGFB
|
platelet-derived growth factor beta polypeptide
|
|
chr9_-_101470836
|
0.027
|
NM_005458
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2
|
|
chr5_+_176560627
|
0.027
|
NM_022455
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr17_+_34058678
|
0.027
|
NM_033315
|
RASL10B
|
RAS-like, family 10, member B
|
|
chr22_-_41032684
|
0.026
|
NM_020831
|
MKL1
|
megakaryoblastic leukemia (translocation) 1
|
|
chr3_+_183873253
|
0.026
|
NM_004423
|
DVL3
|
dishevelled, dsh homolog 3 (Drosophila)
|
|
chr16_-_3068161
|
0.026
|
NM_021195
|
CLDN6
|
claudin 6
|
|
chr19_+_18794391
|
0.026
|
NM_001098482
NM_015321
|
CRTC1
|
CREB regulated transcription coactivator 1
|
|
chr8_+_102504661
|
0.026
|
NM_024915
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr20_+_51588945
|
0.025
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr3_+_189507448
|
0.025
|
NM_001114980
NM_001114981
NM_001114982
|
TP63
|
tumor protein p63
|
|
chr11_+_118978087
|
0.025
|
NM_014807
|
C2CD2L
|
C2CD2-like
|
|
chr16_+_84853585
|
0.025
|
NM_031476
|
CRISPLD2
|
cysteine-rich secretory protein LCCL domain containing 2
|
|
chr3_+_189349215
|
0.025
|
NM_001114978
NM_001114979
NM_003722
|
TP63
|
tumor protein p63
|
|
chr5_-_139422805
|
0.025
|
NM_001184935
NM_004883
NM_013981
NM_013982
NM_013983
|
NRG2
|
neuregulin 2
|
|
chr22_+_22020272
|
0.023
|
NM_014337
NM_148175
NM_148176
|
PPIL2
|
peptidylprolyl isomerase (cyclophilin)-like 2
|
|
chr7_+_150783771
|
0.021
|
NM_001042535
NM_031946
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3
|
|
chr16_+_1664640
|
0.021
|
NM_020825
|
CRAMP1L
|
Crm, cramped-like (Drosophila)
|
|
chr1_-_179112178
|
0.021
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr20_+_51801821
|
0.020
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr11_-_64546201
|
0.020
|
NM_004630
NM_201995
NM_201997
NM_201998
|
SF1
|
splicing factor 1
|
|
chr9_+_140033588
|
0.019
|
NM_000832
NM_001185090
NM_001185091
NM_007327
NM_021569
|
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1
|
|
chr12_-_111180730
|
0.018
|
NM_001244974
NM_002710
|
PPP1CC
|
protein phosphatase 1, catalytic subunit, gamma isozyme
|
|
chr2_+_206547131
|
0.017
|
NM_003872
NM_018534
NM_201264
NM_201266
NM_201267
NM_201279
|
NRP2
|
neuropilin 2
|
|
chr11_+_66036027
|
0.017
|
NM_030981
|
RAB1B
|
RAB1B, member RAS oncogene family
|
|
chr19_+_10764896
|
0.015
|
NM_001137673
NM_004516
NM_012218
NM_017620
NM_153464
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr8_-_145550536
|
0.015
|
NM_012079
|
DGAT1
|
diacylglycerol O-acyltransferase 1
|
|
chr1_-_179198814
|
0.015
|
NM_001136001
NM_001168236
NM_001168237
NM_001168238
NM_007314
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr10_+_80828776
|
0.014
|
NM_020338
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr18_-_67872897
|
0.014
|
NM_173630
|
RTTN
|
rotatin
|
|
chr1_-_21995793
|
0.013
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr1_+_22778343
|
0.013
|
NM_001083621
NM_014870
|
ZBTB40
|
zinc finger and BTB domain containing 40
|
|
chr9_+_132388404
|
0.013
|
NM_014064
|
METTL11A
|
methyltransferase like 11A
|
|
chr17_+_2699731
|
0.012
|
NM_001100398
NM_015085
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr4_-_1242882
|
0.012
|
NM_001012614
NM_001328
|
CTBP1
|
C-terminal binding protein 1
|
|
chr8_+_55047780
|
0.011
|
NM_014175
|
MRPL15
|
mitochondrial ribosomal protein L15
|
|
chr22_+_21771661
|
0.010
|
NM_015094
|
HIC2
|
hypermethylated in cancer 2
|
|
chr2_-_86333277
|
0.009
|
NM_015425
|
POLR1A
|
polymerase (RNA) I polypeptide A, 194kDa
|
|
chr15_-_79298001
|
0.009
|
NM_153815
|
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1
|
|
chr2_+_131513782
|
0.009
|
NM_001105194
NM_001105195
|
FAM123C
|
family with sequence similarity 123C
|
|
chr1_-_21978287
|
0.008
|
NM_001145658
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chrX_-_53713657
|
0.007
|
NM_031407
|
HUWE1
|
HECT, UBA and WWE domain containing 1
|
|
chr5_-_73937248
|
0.007
|
NM_003633
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr10_+_124913759
|
0.005
|
NM_001007793
NM_004725
|
BUB3
|
budding uninhibited by benzimidazoles 3 homolog (yeast)
|
|
chr10_-_101190201
|
0.004
|
NM_002079
|
GOT1
|
glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1)
|
|
chr19_+_56124958
|
0.003
|
NM_001195605
|
ZNF865
|
zinc finger protein 865
|
|
chr11_+_117049838
|
0.002
|
NM_001040455
|
SIDT2
|
SID1 transmembrane family, member 2
|
|
chr19_-_51220194
|
0.002
|
NM_016148
|
SHANK1
|
SH3 and multiple ankyrin repeat domains 1
|