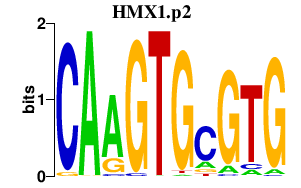
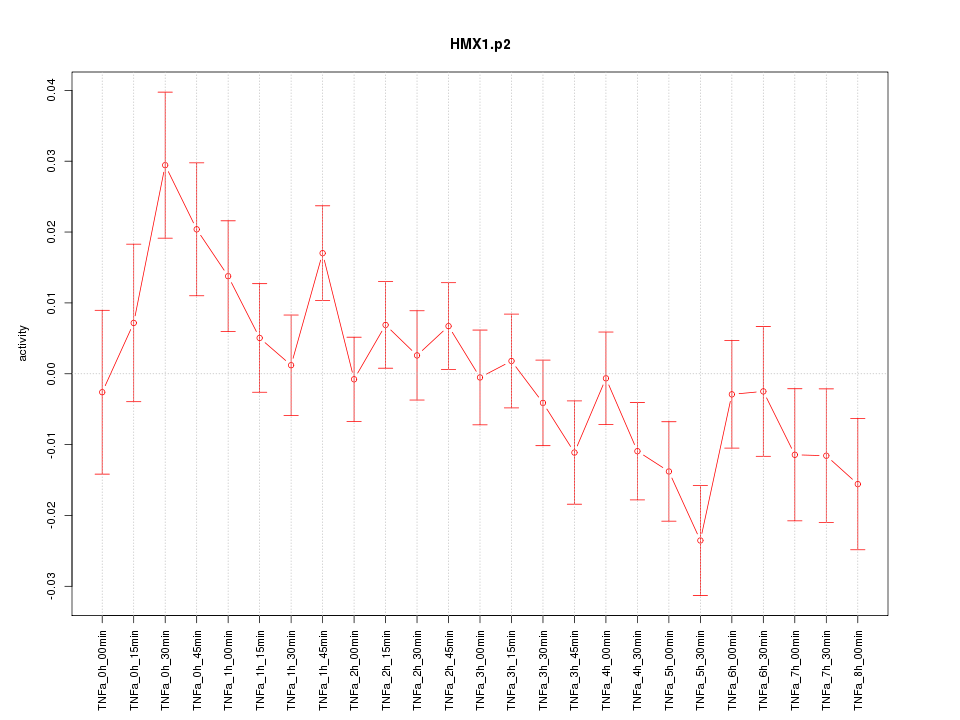
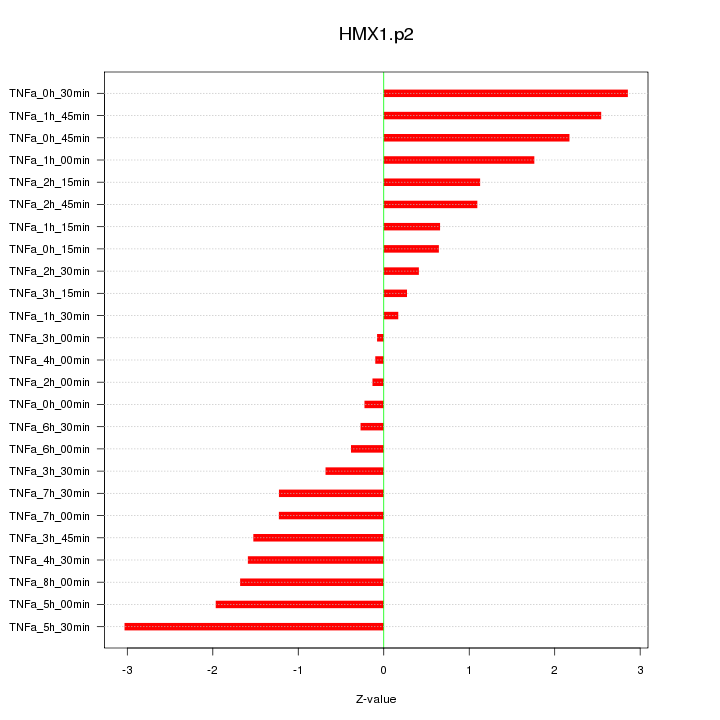
|
chr14_-_105634708
|
6.500
|
|
JAG2
|
jagged 2
|
|
chr5_-_81046858
|
4.264
|
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr1_+_25943958
|
4.209
|
NM_020379
|
MAN1C1
|
mannosidase, alpha, class 1C, member 1
|
|
chr5_+_68788387
|
4.100
|
NM_001205254
|
OCLN
|
occludin
|
|
chr12_+_53440809
|
3.653
|
NM_198316
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr3_-_18466759
|
3.425
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr1_+_221053000
|
3.396
|
|
HLX
|
H2.0-like homeobox
|
|
chr1_-_59042827
|
3.332
|
|
TACSTD2
|
tumor-associated calcium signal transducer 2
|
|
chr5_-_81046943
|
3.305
|
NM_012446
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr6_-_13486986
|
3.110
|
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr14_-_105635114
|
3.079
|
NM_002226
NM_145159
|
JAG2
|
jagged 2
|
|
chr11_-_33890931
|
2.971
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr1_+_221052845
|
2.930
|
|
HLX
|
H2.0-like homeobox
|
|
chr2_-_220436267
|
2.856
|
NM_001173408
NM_001173431
NM_015311
|
OBSL1
|
obscurin-like 1
|
|
chr11_-_33891258
|
2.854
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr2_+_203499819
|
2.691
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr13_+_58207858
|
2.473
|
|
PCDH17
|
protocadherin 17
|
|
chr4_-_186733372
|
2.302
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_-_130031278
|
2.283
|
NM_033515
|
ARHGAP18
|
Rho GTPase activating protein 18
|
|
chr8_-_24772229
|
2.282
|
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr3_-_15900928
|
2.265
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr18_+_72998258
|
2.243
|
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr8_+_37654833
|
2.243
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr6_-_41909386
|
2.234
|
NM_001136125
NM_001760
|
CCND3
|
cyclin D3
|
|
chr11_-_19262424
|
2.122
|
NM_024680
|
E2F8
|
E2F transcription factor 8
|
|
chr1_-_92351613
|
2.120
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr1_+_82266081
|
2.112
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr2_+_112656200
|
2.040
|
|
MERTK
|
c-mer proto-oncogene tyrosine kinase
|
|
chr2_+_112656180
|
1.973
|
NM_006343
|
MERTK
|
c-mer proto-oncogene tyrosine kinase
|
|
chr18_-_45935627
|
1.970
|
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr1_-_27286875
|
1.960
|
NM_152365
|
C1orf172
|
chromosome 1 open reading frame 172
|
|
chr1_-_33168356
|
1.947
|
NM_001161708
NM_030786
|
SYNC
|
syncoilin, intermediate filament protein
|
|
chr12_+_57482674
|
1.938
|
NM_005967
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2)
|
|
chr6_+_16238780
|
1.928
|
NM_006877
|
GMPR
|
guanosine monophosphate reductase
|
|
chr7_-_132261268
|
1.893
|
NM_001105543
NM_020911
|
PLXNA4
|
plexin A4
|
|
chr7_+_22766765
|
1.847
|
NM_000600
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr11_+_59522905
|
1.824
|
|
STX3
|
syntaxin 3
|
|
chr8_+_120220554
|
1.772
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr2_-_220436010
|
1.749
|
|
OBSL1
|
obscurin-like 1
|
|
chr18_+_11981421
|
1.734
|
NM_014214
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2
|
|
chr14_+_63671101
|
1.699
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr4_+_156587861
|
1.691
|
NM_000856
NM_001130682
NM_001130686
NM_001130687
NM_001130683
NM_001130685
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr8_-_93115453
|
1.681
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr12_+_82752275
|
1.678
|
NM_032230
|
C12orf26
|
chromosome 12 open reading frame 26
|
|
chr12_+_57482922
|
1.664
|
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2)
|
|
chr8_+_144816287
|
1.664
|
|
LOC100128338
|
uncharacterized LOC100128338
|
|
chr9_-_130616592
|
1.664
|
|
ENG
|
endoglin
|
|
chr12_-_96184346
|
1.661
|
|
NTN4
|
netrin 4
|
|
chr1_+_25944610
|
1.650
|
|
MAN1C1
|
mannosidase, alpha, class 1C, member 1
|
|
chr12_-_59313304
|
1.628
|
NM_001136051
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr2_-_165478357
|
1.605
|
NM_004490
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr19_+_1026608
|
1.586
|
|
CNN2
|
calponin 2
|
|
chr21_-_40032563
|
1.583
|
NM_001243428
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr18_-_500583
|
1.561
|
NM_130386
|
COLEC12
|
collectin sub-family member 12
|
|
chr12_+_57482886
|
1.537
|
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2)
|
|
chr5_+_139027907
|
1.536
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr7_-_21985421
|
1.533
|
NM_001127370
NM_001127371
NM_018719
|
CDCA7L
|
cell division cycle associated 7-like
|
|
chr10_+_99258615
|
1.531
|
NM_024954
|
UBTD1
|
ubiquitin domain containing 1
|
|
chr20_-_33735060
|
1.526
|
NM_001145025
NM_018217
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2
|
|
chr6_-_2245655
|
1.524
|
NM_001500
|
GMDS
|
GDP-mannose 4,6-dehydratase
|
|
chr9_-_130616760
|
1.521
|
|
ENG
|
endoglin
|
|
chr18_+_11981552
|
1.508
|
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2
|
|
chr20_-_33734894
|
1.507
|
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2
|
|
chr12_-_29936685
|
1.482
|
NM_001193451
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr5_+_68710938
|
1.474
|
NM_001038603
NM_001244734
|
MARVELD2
|
MARVEL domain containing 2
|
|
chr9_-_130617009
|
1.465
|
NM_000118
NM_001114753
|
ENG
|
endoglin
|
|
chr6_-_13486382
|
1.451
|
NM_001242628
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr2_-_191878883
|
1.442
|
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa
|
|
chr19_+_38755097
|
1.409
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr17_+_5973640
|
1.409
|
NM_015253
|
WSCD1
|
WSC domain containing 1
|
|
chr1_+_156863522
|
1.396
|
NM_001080471
|
PEAR1
|
platelet endothelial aggregation receptor 1
|
|
chr15_+_75639330
|
1.394
|
NM_024608
|
NEIL1
|
nei endonuclease VIII-like 1 (E. coli)
|
|
chr9_-_130616957
|
1.389
|
|
ENG
|
endoglin
|
|
chr21_-_44845920
|
1.385
|
|
SIK1
|
salt-inducible kinase 1
|
|
chr11_+_7535000
|
1.380
|
NM_003621
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr17_-_7166263
|
1.378
|
NM_001185023
NM_001307
|
CLDN7
|
claudin 7
|
|
chr11_+_59522801
|
1.377
|
|
STX3
|
syntaxin 3
|
|
chr9_+_22447064
|
1.375
|
|
|
|
|
chr2_-_191878908
|
1.366
|
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa
|
|
chr2_-_191878882
|
1.359
|
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa
|
|
chr6_+_86159738
|
1.359
|
|
NT5E
|
5'-nucleotidase, ecto (CD73)
|
|
chr7_+_22767101
|
1.356
|
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr4_+_42399855
|
1.345
|
NM_001080505
|
SHISA3
|
shisa homolog 3 (Xenopus laevis)
|
|
chr9_+_115513050
|
1.345
|
NM_001012994
|
SNX30
|
sorting nexin family member 30
|
|
chr7_-_752742
|
1.335
|
NM_001164761
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta
|
|
chr19_-_14316980
|
1.333
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr2_+_61293038
|
1.332
|
|
KIAA1841
|
KIAA1841
|
|
chr19_+_38755238
|
1.330
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr2_-_191878924
|
1.329
|
NM_007315
NM_139266
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa
|
|
chr5_+_139027868
|
1.326
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr22_+_50781717
|
1.318
|
NM_001242898
NM_001242899
NM_001242900
NM_014678
|
PPP6R2
|
protein phosphatase 6, regulatory subunit 2
|
|
chr2_-_69870705
|
1.317
|
|
AAK1
|
AP2 associated kinase 1
|
|
chr5_-_88179278
|
1.305
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr7_+_12726413
|
1.303
|
NM_001037164
NM_212460
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr2_-_129076045
|
1.296
|
NM_004807
|
HS6ST1
|
heparan sulfate 6-O-sulfotransferase 1
|
|
chr2_+_61292999
|
1.296
|
NM_001129993
|
KIAA1841
|
KIAA1841
|
|
chr9_+_137967401
|
1.289
|
|
OLFM1
|
olfactomedin 1
|
|
chr7_+_12726910
|
1.280
|
NM_001195396
NM_005738
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr6_+_2765561
|
1.279
|
|
WRNIP1
|
Werner helicase interacting protein 1
|
|
chr17_-_62097937
|
1.276
|
NM_001099786
NM_001099787
NM_001099788
NM_001099789
|
ICAM2
|
intercellular adhesion molecule 2
|
|
chr19_+_38755422
|
1.275
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr6_+_34857021
|
1.274
|
NM_015245
|
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A
|
|
chr5_-_88119604
|
1.273
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr4_-_90758126
|
1.271
|
NM_001146054
NM_007308
NM_000345
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr9_-_10612722
|
1.259
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr9_-_125941304
|
1.248
|
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr22_+_51113069
|
1.239
|
NM_001080420
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3
|
|
chr12_-_96184510
|
1.233
|
NM_021229
|
NTN4
|
netrin 4
|
|
chr2_-_165477818
|
1.219
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chrX_-_70473941
|
1.219
|
|
ZMYM3
|
zinc finger, MYM-type 3
|
|
chr3_+_49711751
|
1.218
|
|
APEH
|
N-acylaminoacyl-peptide hydrolase
|
|
chr1_+_150337145
|
1.215
|
|
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2
|
|
chr12_+_51318743
|
1.212
|
|
METTL7A
|
methyltransferase like 7A
|
|
chr12_+_27397077
|
1.203
|
NM_015000
|
STK38L
|
serine/threonine kinase 38 like
|
|
chr10_+_8097350
|
1.203
|
|
GATA3
|
GATA binding protein 3
|
|
chr13_+_42031679
|
1.198
|
|
C13orf15
|
chromosome 13 open reading frame 15
|
|
chr13_+_42031690
|
1.197
|
|
C13orf15
|
chromosome 13 open reading frame 15
|
|
chr1_+_78511588
|
1.184
|
NM_017655
|
GIPC2
|
GIPC PDZ domain containing family, member 2
|
|
chr13_+_42031525
|
1.170
|
NM_014059
|
C13orf15
|
chromosome 13 open reading frame 15
|
|
chr2_-_27712520
|
1.162
|
NM_015662
|
IFT172
|
intraflagellar transport 172 homolog (Chlamydomonas)
|
|
chr8_-_28243941
|
1.161
|
NM_018660
|
ZNF395
|
zinc finger protein 395
|
|
chr1_-_35920081
|
1.154
|
|
KIAA0319L
|
KIAA0319-like
|
|
chr16_-_86542463
|
1.141
|
|
LOC400550
|
uncharacterized LOC400550
|
|
chr8_-_29120571
|
1.140
|
NM_015254
|
KIF13B
|
kinesin family member 13B
|
|
chr19_+_1026627
|
1.134
|
|
CNN2
|
calponin 2
|
|
chr11_-_61129462
|
1.134
|
NM_001161454
NM_153611
|
CYBASC3
|
cytochrome b, ascorbate dependent 3
|
|
chr7_+_30951414
|
1.131
|
NM_198098
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr1_+_150336584
|
1.120
|
|
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2
|
|
chr5_+_139027955
|
1.117
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr22_+_29702950
|
1.089
|
NM_006478
NM_152236
NM_152237
|
GAS2L1
|
growth arrest-specific 2 like 1
|
|
chr2_-_165477986
|
1.088
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr4_-_102267952
|
1.085
|
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr19_+_50796820
|
1.084
|
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chr6_-_7910716
|
1.078
|
NM_030810
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum)
|
|
chrX_+_101967290
|
1.077
|
|
GPRASP2
|
G protein-coupled receptor associated sorting protein 2
|
|
chr17_-_8534008
|
1.077
|
NM_005964
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr3_-_125775609
|
1.072
|
NM_001008487
|
SLC41A3
|
solute carrier family 41, member 3
|
|
chr12_+_3068442
|
1.061
|
NM_003213
NM_201441
NM_201443
|
TEAD4
|
TEA domain family member 4
|
|
chr11_-_116968986
|
1.052
|
NM_025164
|
SIK3
|
SIK family kinase 3
|
|
chr11_+_7618416
|
1.049
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr17_-_34313685
|
1.046
|
NM_032962
NM_032963
|
CCL14
|
chemokine (C-C motif) ligand 14
|
|
chr11_+_68079999
|
1.045
|
NM_002335
|
LRP5
|
low density lipoprotein receptor-related protein 5
|
|
chr4_-_114682836
|
1.042
|
NM_001221
NM_172114
NM_172115
NM_172127
NM_172128
NM_172129
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta
|
|
chr19_+_1026271
|
1.039
|
NM_004368
NM_201277
|
CNN2
|
calponin 2
|
|
chr20_-_62601140
|
1.028
|
NM_020713
|
ZNF512B
|
zinc finger protein 512B
|
|
chr6_+_84743250
|
1.025
|
NM_138409
|
MRAP2
|
melanocortin 2 receptor accessory protein 2
|
|
chr17_+_19551835
|
1.018
|
NM_000382
NM_001031806
|
ALDH3A2
|
aldehyde dehydrogenase 3 family, member A2
|
|
chr15_-_90198524
|
1.007
|
NM_198525
|
KIF7
|
kinesin family member 7
|
|
chr4_-_107237794
|
1.005
|
NM_001163436
|
TBCK
|
TBC1 domain containing kinase
|
|
chr20_-_35374530
|
0.995
|
NM_022477
NM_032013
|
NDRG3
|
NDRG family member 3
|
|
chr10_-_33623771
|
0.991
|
NM_001024628
NM_001024629
NM_001244972
NM_001244973
NM_003873
|
NRP1
|
neuropilin 1
|
|
chr19_-_36545119
|
0.988
|
|
THAP8
|
THAP domain containing 8
|
|
chr11_+_66624501
|
0.987
|
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr6_+_7727010
|
0.978
|
NM_001718
|
BMP6
|
bone morphogenetic protein 6
|
|
chr15_-_82336880
|
0.976
|
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr22_+_50781773
|
0.975
|
|
PPP6R2
|
protein phosphatase 6, regulatory subunit 2
|
|
chr3_+_57994087
|
0.972
|
NM_001164317
NM_001164318
NM_001164319
NM_001457
|
FLNB
|
filamin B, beta
|
|
chr1_+_145524989
|
0.971
|
NM_003637
|
ITGA10
|
integrin, alpha 10
|
|
chr7_+_130131921
|
0.963
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr20_-_35374468
|
0.963
|
|
NDRG3
|
NDRG family member 3
|
|
chr2_-_192015733
|
0.960
|
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr3_-_183735617
|
0.958
|
NM_001023587
NM_005688
|
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5
|
|
chr1_-_24194760
|
0.956
|
NM_000147
|
FUCA1
|
fucosidase, alpha-L- 1, tissue
|
|
chr8_+_143808593
|
0.955
|
NM_016647
|
C8orf55
|
chromosome 8 open reading frame 55
|
|
chr7_-_37488408
|
0.948
|
|
ELMO1
|
engulfment and cell motility 1
|
|
chr7_+_12727204
|
0.944
|
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr19_-_19006868
|
0.934
|
NM_021267
NM_198207
NM_001492
|
CERS1
GDF1
|
ceramide synthase 1
growth differentiation factor 1
|
|
chr12_-_42877785
|
0.932
|
NM_001144882
NM_001144883
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chr16_+_68119211
|
0.925
|
NM_004555
NM_173163
NM_173165
|
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3
|
|
chr1_+_64058891
|
0.915
|
NM_002633
|
PGM1
|
phosphoglucomutase 1
|
|
chr4_-_110650965
|
0.911
|
NM_030821
|
PLA2G12A
|
phospholipase A2, group XIIA
|
|
chr15_+_83776323
|
0.902
|
NM_001144903
NM_023003
|
TM6SF1
|
transmembrane 6 superfamily member 1
|
|
chr2_-_192015751
|
0.897
|
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr9_+_131644427
|
0.893
|
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr7_-_37488554
|
0.886
|
NM_014800
|
ELMO1
|
engulfment and cell motility 1
|
|
chr19_+_18530087
|
0.880
|
NM_001009998
NM_032627
|
SSBP4
|
single stranded DNA binding protein 4
|
|
chr17_-_7165788
|
0.876
|
|
CLDN7
|
claudin 7
|
|
chr3_+_50192420
|
0.876
|
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F
|
|
chr22_+_19744225
|
0.872
|
NM_005992
NM_080646
NM_080647
|
TBX1
|
T-box 1
|
|
chr4_+_156588811
|
0.868
|
NM_001130684
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr1_-_225615701
|
0.867
|
|
LBR
|
lamin B receptor
|
|
chrX_-_153718937
|
0.850
|
|
SLC10A3
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 3
|
|
chr9_+_131644368
|
0.843
|
NM_001127245
NM_019594
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chrX_-_70473979
|
0.842
|
NM_201599
|
ZMYM3
|
zinc finger, MYM-type 3
|
|
chr2_-_192015706
|
0.841
|
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr2_-_192015934
|
0.835
|
NM_003151
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr9_-_85677784
|
0.829
|
|
RASEF
|
RAS and EF-hand domain containing
|
|
chr8_-_28347733
|
0.829
|
NM_172366
|
FBXO16
ZNF395
|
F-box protein 16
zinc finger protein 395
|
|
chr11_+_66624875
|
0.822
|
NM_024036
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr15_-_102029872
|
0.819
|
|
PCSK6
|
proprotein convertase subtilisin/kexin type 6
|
|
chr10_-_52383611
|
0.818
|
NM_147156
|
SGMS1
|
sphingomyelin synthase 1
|
|
chrX_+_150863729
|
0.816
|
NM_024082
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane)
|
|
chr15_-_56035176
|
0.813
|
NM_173814
|
PRTG
|
protogenin
|
|
chrX_-_70474402
|
0.810
|
NM_001171163
NM_005096
|
ZMYM3
|
zinc finger, MYM-type 3
|
|
chr1_+_92495484
|
0.810
|
NM_173567
|
EPHX4
|
epoxide hydrolase 4
|
|
chr1_+_26856259
|
0.810
|
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr8_-_28243589
|
0.802
|
|
ZNF395
|
zinc finger protein 395
|
|
chr6_-_8102753
|
0.801
|
NM_001135650
NM_004280
|
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1
|
|
chrX_-_153718984
|
0.799
|
NM_001142391
NM_001142392
NM_019848
|
SLC10A3
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 3
|
|
chr11_-_119252267
|
0.799
|
|
USP2
|
ubiquitin specific peptidase 2
|