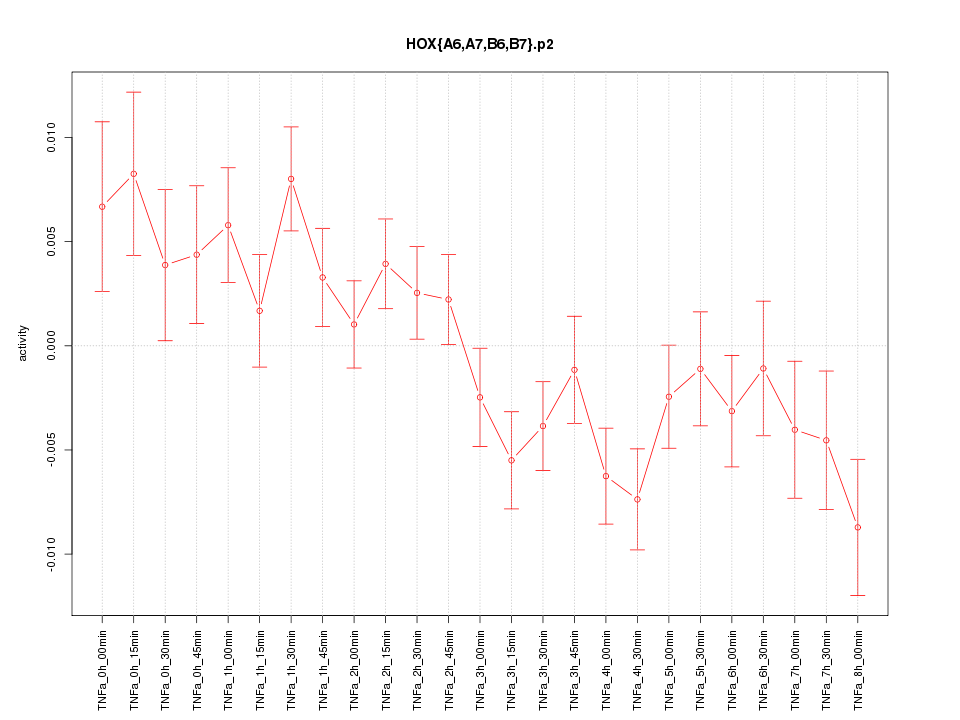
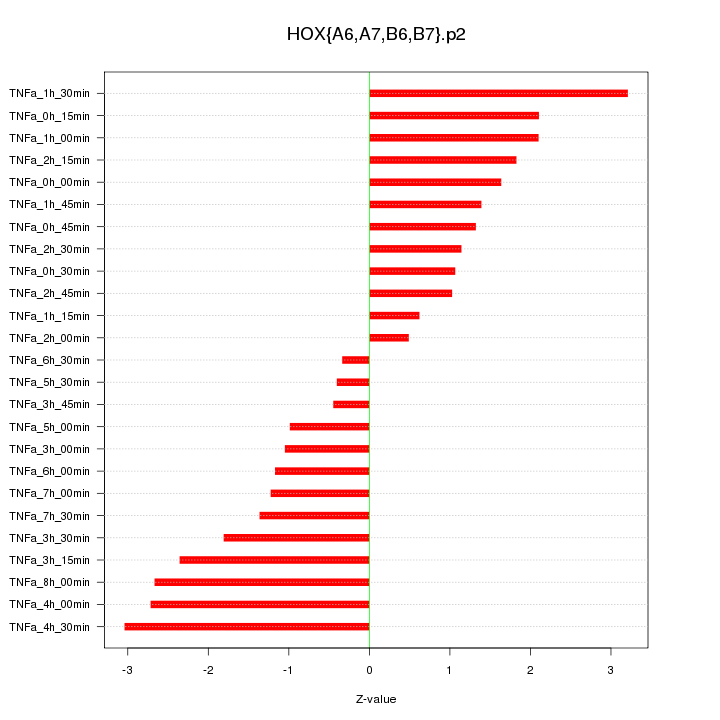
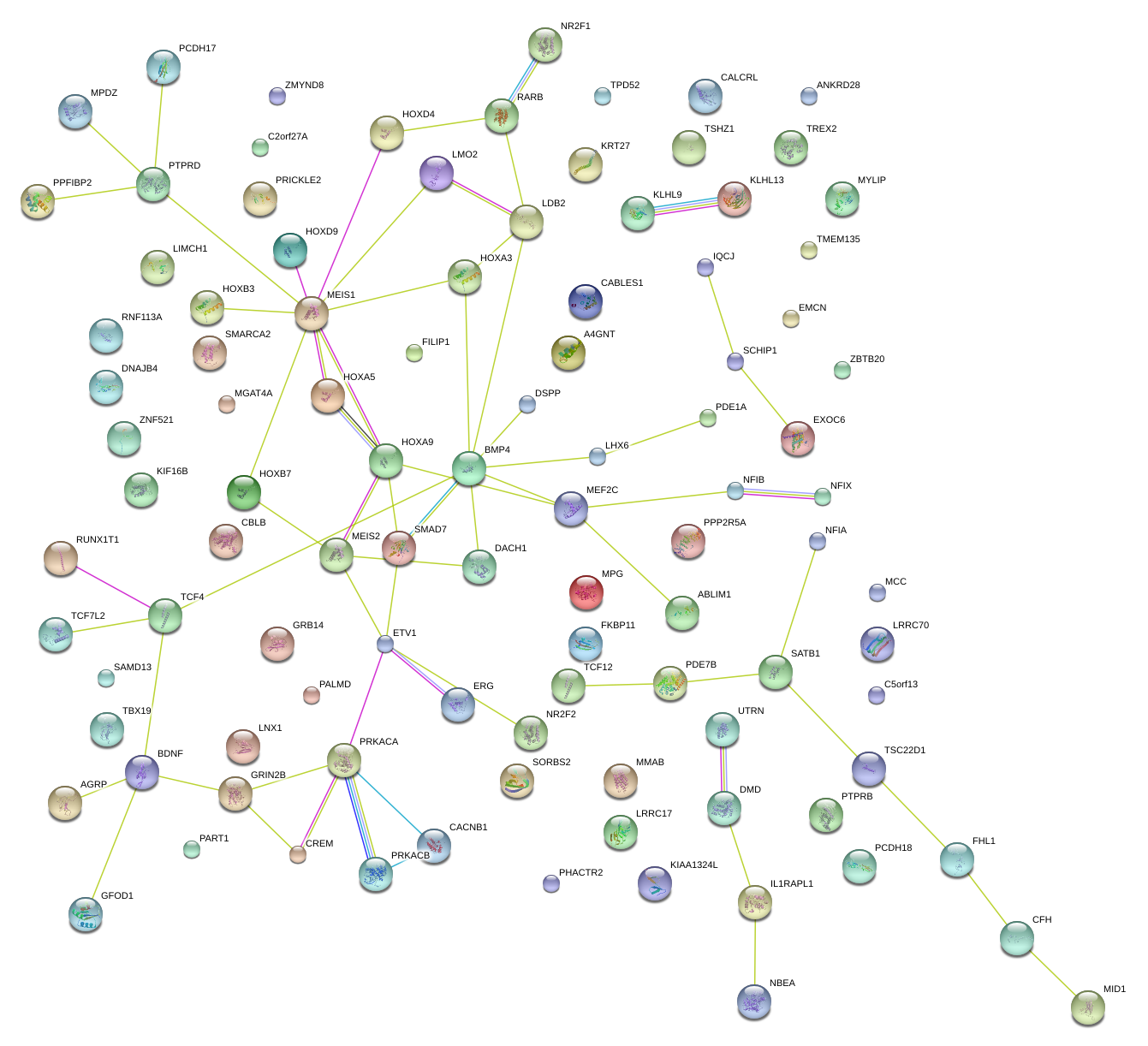
|
chr9_-_14086901
|
31.534
|
|
NFIB
|
nuclear factor I/B
|
|
chr1_+_61869699
|
20.504
|
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_84630575
|
16.138
|
NM_001242860
NM_001242861
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr1_+_84630014
|
13.228
|
NM_001242857
NM_001242858
NM_001242859
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr5_-_88119604
|
13.158
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr4_+_41614725
|
12.921
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr4_+_41614911
|
12.479
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr4_+_41614933
|
12.047
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr5_-_88199868
|
11.429
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr4_-_186877869
|
9.282
|
NM_021069
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_+_41700736
|
8.931
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr3_-_64211111
|
8.079
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr1_+_84630374
|
7.782
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr7_-_27183225
|
7.186
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr8_-_93107442
|
7.175
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_+_61547388
|
6.550
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr14_-_54423448
|
6.538
|
NM_001202
NM_130850
|
BMP4
|
bone morphogenetic protein 4
|
|
chr18_-_53089722
|
6.461
|
NM_001243233
|
TCF4
|
transcription factor 4
|
|
chr1_+_100111430
|
6.451
|
NM_017734
|
PALMD
|
palmdelphin
|
|
chr4_-_186733372
|
6.374
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr18_-_22932109
|
6.317
|
NM_015461
|
ZNF521
|
zinc finger protein 521
|
|
chr5_-_88179278
|
6.133
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr8_-_80993009
|
6.021
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr18_-_22931970
|
5.807
|
|
ZNF521
|
zinc finger protein 521
|
|
chr9_-_8733893
|
5.432
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr6_-_76203256
|
5.416
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr1_+_84767288
|
5.268
|
NM_001134663
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr18_+_20735788
|
5.242
|
NM_138375
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr11_+_87032428
|
5.166
|
|
TMEM135
|
transmembrane protein 135
|
|
chr9_+_2015218
|
5.126
|
NM_003070
NM_139045
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2
|
|
chr6_+_136172802
|
4.913
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr1_+_84647342
|
4.869
|
NM_001242862
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr7_-_27205123
|
4.749
|
|
HOXA9
|
homeobox A9
|
|
chr10_-_116392099
|
4.689
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr7_-_27166638
|
4.630
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr4_-_16900348
|
4.481
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr1_+_212475147
|
4.441
|
NM_001199756
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha
|
|
chr5_-_111091947
|
4.349
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr13_-_72441312
|
4.221
|
NM_004392
NM_080759
NM_080760
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr10_-_116418056
|
4.219
|
NM_002313
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr8_-_93107643
|
4.205
|
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr18_+_72922725
|
4.197
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr1_+_78470593
|
4.093
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr2_-_183387063
|
4.082
|
NM_005019
NM_001003683
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr1_+_84609941
|
4.019
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr17_-_46687925
|
3.995
|
|
HOXB7
|
homeobox B7
|
|
chr7_-_27205148
|
3.970
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr3_-_15839674
|
3.938
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chrX_-_117119282
|
3.870
|
NM_001168299
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr1_+_61547625
|
3.808
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr3_+_25470067
|
3.709
|
|
RARB
|
retinoic acid receptor, beta
|
|
chr10_+_35426709
|
3.701
|
NM_183011
NM_183012
|
CREM
|
cAMP responsive element modulator
|
|
chr15_+_57511616
|
3.683
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr20_-_45976626
|
3.648
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr2_-_165424993
|
3.635
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr2_-_188312872
|
3.580
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chrX_+_135251795
|
3.562
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr2_+_66662692
|
3.553
|
|
MEIS1
|
Meis homeobox 1
|
|
chr7_-_14025826
|
3.405
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr13_+_58205943
|
3.384
|
|
PCDH17
|
protocadherin 17
|
|
chr8_-_93107881
|
3.355
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr13_+_36050885
|
3.352
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr10_+_35456443
|
3.247
|
NM_182771
NM_182772
|
CREM
|
cAMP responsive element modulator
|
|
chr11_-_27742224
|
3.155
|
NM_001143805
NM_001143806
NM_170732
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr6_+_143929316
|
3.150
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr18_-_46469118
|
3.121
|
NM_001190823
|
SMAD7
|
SMAD family member 7
|
|
chr21_-_39870303
|
3.078
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr11_+_7618416
|
3.076
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr17_-_46688279
|
3.041
|
|
HOXB7
|
homeobox B7
|
|
chr2_+_66662516
|
2.944
|
NM_002398
|
MEIS1
|
Meis homeobox 1
|
|
chr3_-_18480203
|
2.916
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr6_+_16143281
|
2.903
|
|
MYLIP
|
myosin regulatory light chain interacting protein
|
|
chr9_-_14398981
|
2.888
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr6_+_144612872
|
2.874
|
NM_007124
|
UTRN
|
utrophin
|
|
chr10_+_94594469
|
2.858
|
NM_001013848
|
EXOC6
|
exocyst complex component 6
|
|
chrX_+_135278912
|
2.850
|
NM_001159701
NM_001159699
|
FHL1
|
four and a half LIM domains 1
|
|
chr5_+_59784004
|
2.814
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr1_+_168250277
|
2.808
|
NM_005149
|
TBX19
|
T-box 19
|
|
chr5_+_61874561
|
2.791
|
NM_181506
|
LRRC70
|
leucine rich repeat containing 70
|
|
chr17_-_46688362
|
2.786
|
NM_004502
|
HOXB7
|
homeobox B7
|
|
chr12_-_71031174
|
2.776
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr4_-_16900028
|
2.753
|
|
LDB2
|
LIM domain binding 2
|
|
chr2_+_132480063
|
2.749
|
NM_013310
|
C2orf27A
|
chromosome 2 open reading frame 27A
|
|
chr6_-_13408368
|
2.749
|
NM_001242630
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr12_-_14132920
|
2.739
|
NM_000834
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B
|
|
chr4_-_16900256
|
2.730
|
|
LDB2
|
LIM domain binding 2
|
|
chrX_+_28605558
|
2.725
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr17_-_46651809
|
2.718
|
NM_002146
|
HOXB3
|
homeobox B3
|
|
chr20_-_45985411
|
2.705
|
NM_012408
NM_183047
NM_183048
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr2_+_177016112
|
2.675
|
NM_014621
|
HOXD4
|
homeobox D4
|
|
chr13_+_58205788
|
2.652
|
NM_001040429
|
PCDH17
|
protocadherin 17
|
|
chr9_-_13250313
|
2.616
|
NM_003829
|
MPDZ
|
multiple PDZ domain protein
|
|
chr13_-_45010937
|
2.608
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr3_-_114478117
|
2.601
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr15_+_96869156
|
2.562
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chrX_-_117107700
|
2.560
|
NM_033495
NM_001168300
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chrX_+_135251454
|
2.539
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr4_-_101439071
|
2.470
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr18_-_53071225
|
2.466
|
NM_001243232
|
TCF4
|
transcription factor 4
|
|
chr20_-_45927496
|
2.442
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr3_-_137851123
|
2.393
|
NM_016161
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase
|
|
chr2_-_99279935
|
2.391
|
NM_001160154
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr4_+_88529680
|
2.389
|
NM_014208
|
DSPP
|
dentin sialophosphoprotein
|
|
chr4_-_138453628
|
2.387
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr2_+_177016337
|
2.365
|
|
|
|
|
chr3_-_105421044
|
2.345
|
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr11_-_33913835
|
2.319
|
NM_005574
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr18_-_52988916
|
2.316
|
NM_001243234
NM_001243235
|
TCF4
|
transcription factor 4
|
|
chrX_-_32430370
|
2.256
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr5_-_168271852
|
2.254
|
|
|
|
|
chr7_-_86595203
|
2.227
|
NM_152748
|
KIAA1324L
|
KIAA1324-like
|
|
chrX_-_33357725
|
2.210
|
NM_000109
|
DMD
|
dystrophin
|
|
chr5_-_112630557
|
2.158
|
NM_002387
|
MCC
|
mutated in colorectal cancers
|
|
chr10_-_116444374
|
2.149
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr8_-_93029864
|
2.144
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr16_-_67517715
|
2.133
|
NM_001138
|
AGRP
|
agouti related protein homolog (mouse)
|
|
chr3_-_18466759
|
2.113
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr7_+_102553343
|
2.108
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chrX_-_10535476
|
2.101
|
NM_001193278
NM_001193279
NM_001193280
NM_001193281
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr17_-_38938785
|
2.082
|
NM_181537
|
KRT27
|
keratin 27
|
|
chr9_-_124989755
|
2.060
|
NM_001242334
|
LHX6
|
LIM homeobox 6
|
|
chr3_+_159557745
|
2.035
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr10_+_83637442
|
2.019
|
NM_001165973
|
NRG3
|
neuregulin 3
|
|
chr1_+_100326765
|
2.010
|
NM_000645
|
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase
|
|
chr1_+_244214576
|
1.995
|
|
ZNF238
|
zinc finger protein 238
|
|
chr2_+_33359607
|
1.985
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr4_-_186877413
|
1.975
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr11_-_27741192
|
1.959
|
NM_001143807
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr9_-_14314036
|
1.959
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr18_+_42260774
|
1.956
|
NM_015559
|
SETBP1
|
SET binding protein 1
|
|
chr12_+_32654976
|
1.952
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chrX_-_19817854
|
1.945
|
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr18_-_53177999
|
1.944
|
NM_001243231
|
TCF4
|
transcription factor 4
|
|
chr10_-_126716452
|
1.928
|
NM_022802
|
CTBP2
|
C-terminal binding protein 2
|
|
chr7_-_27213873
|
1.923
|
NM_018951
|
HOXA10
|
homeobox A10
|
|
chr4_-_138453564
|
1.916
|
|
PCDH18
|
protocadherin 18
|
|
chr14_+_38678280
|
1.907
|
|
SSTR1
|
somatostatin receptor 1
|
|
chr17_-_1248346
|
1.903
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr11_-_27723061
|
1.900
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chrX_-_32173585
|
1.891
|
NM_004013
NM_004020
NM_004021
NM_004022
NM_004023
|
DMD
|
dystrophin
|
|
chr10_+_35464514
|
1.880
|
NM_182769
NM_182770
|
CREM
|
cAMP responsive element modulator
|
|
chr1_-_76397975
|
1.873
|
NM_080868
|
ASB17
|
ankyrin repeat and SOCS box containing 17
|
|
chr12_+_14538232
|
1.869
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr11_+_58390145
|
1.850
|
NM_000614
|
CNTF
|
ciliary neurotrophic factor
|
|
chr6_+_153019029
|
1.843
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chr12_-_50616425
|
1.841
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr20_+_11898536
|
1.835
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr13_-_48987652
|
1.822
|
NM_001162498
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr4_+_140587086
|
1.821
|
|
MGST2
|
microsomal glutathione S-transferase 2
|
|
chr5_+_42565963
|
1.819
|
NM_001242460
NM_001242461
NM_001242462
|
GHR
|
growth hormone receptor
|
|
chr14_-_92413738
|
1.818
|
|
FBLN5
|
fibulin 5
|
|
chr4_-_186577858
|
1.816
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr11_-_71752099
|
1.809
|
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr9_+_79115548
|
1.806
|
NM_001097636
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr4_+_170581212
|
1.800
|
NM_001243374
|
CLCN3
|
chloride channel 3
|
|
chr3_+_113616317
|
1.796
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr16_+_53133063
|
1.766
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr5_-_39394423
|
1.757
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chrX_-_18690187
|
1.753
|
NM_000330
|
RS1
|
retinoschisin 1
|
|
chr4_-_186578122
|
1.744
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr10_-_33623299
|
1.722
|
|
NRP1
|
neuropilin 1
|
|
chr4_-_141075232
|
1.721
|
NM_018717
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr3_+_132318980
|
1.721
|
NM_178445
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr21_-_40033617
|
1.715
|
NM_001243432
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr1_-_94586678
|
1.715
|
NM_000350
|
ABCA4
|
ATP-binding cassette, sub-family A (ABC1), member 4
|
|
chr15_+_96876568
|
1.695
|
NM_001145157
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr9_+_97521930
|
1.695
|
NM_001193329
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr9_-_113100076
|
1.692
|
NM_001003936
|
TXNDC8
|
thioredoxin domain containing 8 (spermatozoa)
|
|
chrX_+_102585113
|
1.682
|
NM_152278
|
TCEAL7
|
transcription elongation factor A (SII)-like 7
|
|
chr2_-_177502658
|
1.680
|
|
LOC375295
|
uncharacterized LOC375295
|
|
chr8_-_17752847
|
1.679
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr12_+_80603232
|
1.679
|
NM_173591
|
OTOGL
|
otogelin-like
|
|
chr1_+_244216506
|
1.678
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr2_-_198540583
|
1.678
|
NM_144629
|
RFTN2
|
raftlin family member 2
|
|
chr10_+_63808968
|
1.671
|
NM_001244638
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr3_-_168864066
|
1.669
|
NM_001105077
NM_001163999
NM_005241
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr4_-_159093523
|
1.665
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr5_+_175108463
|
1.648
|
NM_022304
|
HRH2
|
histamine receptor H2
|
|
chr20_+_42574535
|
1.631
|
NM_001098798
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr12_+_79258588
|
1.624
|
|
SYT1
|
synaptotagmin I
|
|
chr5_+_139505519
|
1.618
|
NM_001007189
|
IGIP
|
IgA-inducing protein homolog (Bos taurus)
|
|
chr5_-_90679115
|
1.609
|
NM_020801
|
ARRDC3
|
arrestin domain containing 3
|
|
chr17_-_16097882
|
1.607
|
NM_001190438
NM_001190440
|
NCOR1
|
nuclear receptor corepressor 1
|
|
chr10_-_64576074
|
1.593
|
|
EGR2
|
early growth response 2
|
|
chr8_-_93111915
|
1.590
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr4_-_146859960
|
1.566
|
|
|
|
|
chr5_+_95066628
|
1.558
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|
|
chr8_-_93075190
|
1.546
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr6_-_32157511
|
1.539
|
NM_002586
|
PBX2
|
pre-B-cell leukemia homeobox 2
|
|
chr10_+_112344076
|
1.535
|
|
SMC3
|
structural maintenance of chromosomes 3
|
|
chr3_-_114343052
|
1.534
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr6_+_87969619
|
1.503
|
|
ZNF292
|
zinc finger protein 292
|
|
chr4_-_2230957
|
1.500
|
NM_181808
|
POLN
|
polymerase (DNA directed) nu
|
|
chr14_+_70236266
|
1.496
|
|
SRSF5
|
serine/arginine-rich splicing factor 5
|
|
chr2_-_225266710
|
1.488
|
NM_001122779
NM_024785
|
FAM124B
|
family with sequence similarity 124B
|
|
chr18_+_33234532
|
1.488
|
NM_020474
|
GALNT1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1)
|
|
chr3_+_148545587
|
1.488
|
NM_001871
|
CPB1
|
carboxypeptidase B1 (tissue)
|
|
chr10_-_64576123
|
1.478
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr17_-_35656691
|
1.476
|
NM_198837
NM_198838
|
ACACA
|
acetyl-CoA carboxylase alpha
|
|
chr1_+_33004771
|
1.470
|
NM_001040441
|
ZBTB8A
|
zinc finger and BTB domain containing 8A
|