|
chr4_+_90033967
|
0.914
|
NM_145715
|
TIGD2
|
tigger transposable element derived 2
|
|
chrX_+_65382432
|
0.901
|
NM_138737
|
HEPH
|
hephaestin
|
|
chr3_-_18480203
|
0.830
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr7_-_27183225
|
0.809
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr1_-_76397975
|
0.781
|
NM_080868
|
ASB17
|
ankyrin repeat and SOCS box containing 17
|
|
chr1_+_84768333
|
0.762
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr17_-_48945338
|
0.761
|
NM_001243877
|
TOB1
|
transducer of ERBB2, 1
|
|
chr7_-_27159213
|
0.471
|
NM_030661
|
HOXA3
|
homeobox A3
|
|
chr7_+_100026412
|
0.458
|
NM_001194990
NM_001194991
NM_001194992
|
MEPCE
|
methylphosphate capping enzyme
|
|
chr4_+_41614911
|
0.434
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr8_-_93075190
|
0.425
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr10_+_97889467
|
0.418
|
NM_014803
|
ZNF518A
|
zinc finger protein 518A
|
|
chr7_+_100027253
|
0.418
|
NM_019606
|
MEPCE
|
methylphosphate capping enzyme
|
|
chr18_+_47088400
|
0.415
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr19_+_39899749
|
0.414
|
|
|
|
|
chr3_-_74570262
|
0.397
|
NM_020872
|
CNTN3
|
contactin 3 (plasmacytoma associated)
|
|
chr6_-_26108320
|
0.394
|
NM_005323
|
HIST1H1T
|
histone cluster 1, H1t
|
|
chr22_+_22735202
|
0.370
|
|
|
|
|
chr8_-_93111915
|
0.364
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr12_-_14132920
|
0.338
|
NM_000834
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B
|
|
chr3_+_127641901
|
0.335
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr11_-_27681195
|
0.332
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr7_+_100026413
|
0.319
|
|
MEPCE
|
methylphosphate capping enzyme
|
|
chr4_+_41614725
|
0.313
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr8_-_93107881
|
0.302
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chrX_+_65382627
|
0.292
|
|
HEPH
|
hephaestin
|
|
chr5_+_139505519
|
0.289
|
NM_001007189
|
IGIP
|
IgA-inducing protein homolog (Bos taurus)
|
|
chr17_-_67240955
|
0.286
|
NM_080282
|
ABCA10
|
ATP-binding cassette, sub-family A (ABC1), member 10
|
|
chr13_-_52980628
|
0.283
|
NM_018676
NM_199263
|
THSD1
|
thrombospondin, type I, domain containing 1
|
|
chr6_+_88182679
|
0.274
|
|
SLC35A1
|
solute carrier family 35 (CMP-sialic acid transporter), member A1
|
|
chr19_-_22379752
|
0.272
|
NM_001001411
|
ZNF676
|
zinc finger protein 676
|
|
chr11_+_5509914
|
0.264
|
NM_001005163
|
OR52D1
|
olfactory receptor, family 52, subfamily D, member 1
|
|
chr13_-_45010974
|
0.260
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr4_+_41614933
|
0.256
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr6_+_88182624
|
0.254
|
NM_001168398
NM_006416
|
SLC35A1
|
solute carrier family 35 (CMP-sialic acid transporter), member A1
|
|
chr1_-_116311161
|
0.250
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr13_-_45010937
|
0.250
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr5_+_140602914
|
0.248
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr13_-_45010994
|
0.244
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr1_-_116311330
|
0.240
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr6_-_130031278
|
0.235
|
NM_033515
|
ARHGAP18
|
Rho GTPase activating protein 18
|
|
chr1_-_116311398
|
0.235
|
NM_001232
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr18_+_8717368
|
0.233
|
NM_015210
|
CCDC165
|
coiled-coil domain containing 165
|
|
chr22_-_22863504
|
0.232
|
NM_080764
|
ZNF280B
|
zinc finger protein 280B
|
|
chr1_+_84647342
|
0.228
|
NM_001242862
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr10_+_94594469
|
0.220
|
NM_001013848
|
EXOC6
|
exocyst complex component 6
|
|
chr1_+_61869699
|
0.217
|
|
NFIA
|
nuclear factor I/A
|
|
chr2_-_158731622
|
0.211
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr12_-_11150473
|
0.210
|
NM_176889
|
TAS2R20
|
taste receptor, type 2, member 20
|
|
chr6_+_144612872
|
0.208
|
NM_007124
|
UTRN
|
utrophin
|
|
chr19_-_48759141
|
0.204
|
NM_001184902
NM_001184904
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr3_+_46919235
|
0.204
|
NM_000316
|
PTH1R
|
parathyroid hormone 1 receptor
|
|
chr12_-_11214892
|
0.202
|
NM_176887
|
TAS2R46
|
taste receptor, type 2, member 46
|
|
chr4_+_36285643
|
0.201
|
NM_001170700
|
DTHD1
|
death domain containing 1
|
|
chr3_+_97868169
|
0.201
|
NM_001005514
|
OR5H14
|
olfactory receptor, family 5, subfamily H, member 14
|
|
chr9_+_19290748
|
0.200
|
NM_017925
|
DENND4C
|
DENN/MADD domain containing 4C
|
|
chr12_-_51611476
|
0.198
|
|
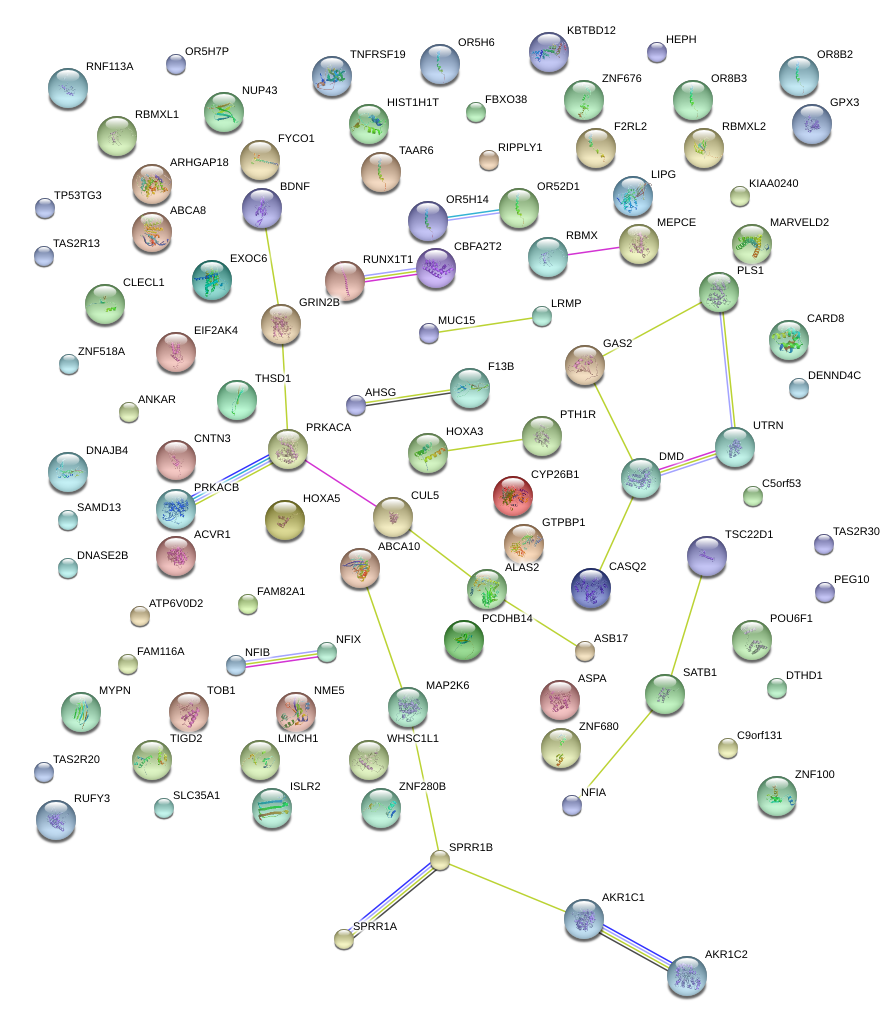
POU6F1
|
POU class 6 homeobox 1
|
|
chrX_-_106146546
|
0.194
|
NM_001171706
NM_138382
|
RIPPLY1
|
ripply1 homolog (zebrafish)
|
|
chrX_-_135962872
|
0.191
|
|
RBMX
|
RNA binding motif protein, X-linked
|
|
chr10_+_69869249
|
0.188
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr1_+_15272414
|
0.183
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr10_+_5005685
|
0.180
|
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr11_+_22696318
|
0.178
|
NM_005256
|
GAS2
|
growth arrest-specific 2
|
|
chr1_+_78470593
|
0.174
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr5_-_137475076
|
0.171
|
NM_003551
|
NME5
|
non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr11_+_107879407
|
0.168
|
NM_003478
|
CUL5
|
cullin 5
|
|
chr5_+_147795459
|
0.162
|
|
FBXO38
|
F-box protein 38
|
|
chr1_+_153003677
|
0.158
|
NM_003125
|
SPRR1B
|
small proline-rich protein 1B
|
|
chr2_+_190540710
|
0.157
|
NM_144708
|
ANKAR
|
ankyrin and armadillo repeat containing
|
|
chr12_-_9885859
|
0.152
|
NM_001253748
NM_001253750
NM_172004
|
CLECL1
|
C-type lectin-like 1
|
|
chrX_-_33229428
|
0.143
|
NM_004006
|
DMD
|
dystrophin
|
|
chr10_-_5045967
|
0.143
|
NM_001135241
|
AKR1C2
|
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chr11_-_124253238
|
0.140
|
NM_001005468
|
OR8B2
|
olfactory receptor, family 8, subfamily B, member 2
|
|
chr16_-_32688052
|
0.140
|
NM_016212
|
TP53TG3
|
TP53 target 3
|
|
chr2_-_72371188
|
0.139
|
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chr16_+_33261514
|
0.138
|
NM_016212
|
TP53TG3
|
TP53 target 3
|
|
chr2_+_38155901
|
0.137
|
NM_001170792
|
FAM82A1
|
family with sequence similarity 82, member A1
|
|
chr7_+_94297448
|
0.136
|
|
PEG10
|
paternally expressed 10
|
|
chr11_-_26593667
|
0.134
|
NM_001135091
NM_001135092
NM_145650
|
MUC15
|
mucin 15, cell surface associated
|
|
chr12_-_11062160
|
0.133
|
NM_023920
|
TAS2R13
|
taste receptor, type 2, member 13
|
|
chr22_+_39101954
|
0.132
|
|
GTPBP1
|
GTP binding protein 1
|
|
chr12_+_25205488
|
0.130
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr10_+_5005453
|
0.129
|
NM_001353
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr15_+_40294749
|
0.125
|
|
EIF2AK4
|
eukaryotic translation initiation factor 2 alpha kinase 4
|
|
chr9_+_35041101
|
0.121
|
NM_001040411
NM_001040412
|
C9orf131
|
chromosome 9 open reading frame 131
|
|
chr17_+_3379295
|
0.120
|
NM_000049
|
ASPA
|
aspartoacylase
|
|
chr3_-_13057130
|
0.120
|
|
|
|
|
chr19_-_48752921
|
0.118
|
NM_001184901
NM_001184903
NM_014959
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr7_-_64023454
|
0.117
|
NM_001130022
NM_178558
|
ZNF680
|
zinc finger protein 680
|
|
chr16_+_33204979
|
0.116
|
NM_016212
|
TP53TG3
|
TP53 target 3
|
|
chr15_+_74422920
|
0.114
|
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr8_+_87111132
|
0.113
|
NM_152565
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2
|
|
chr1_+_84874033
|
0.113
|
NM_058248
|
DNASE2B
|
deoxyribonuclease II beta
|
|
chr13_+_24144402
|
0.112
|
NM_001204459
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr5_-_75919051
|
0.110
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr5_+_68710938
|
0.109
|
NM_001038603
NM_001244734
|
MARVELD2
|
MARVEL domain containing 2
|
|
chrX_-_55057408
|
0.109
|
NM_000032
NM_001037967
NM_001037968
|
ALAS2
|
aminolevulinate, delta-, synthase 2
|
|
chr4_+_71587674
|
0.105
|
NM_001037442
NM_014961
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chr11_+_22689653
|
0.104
|
NM_177553
|
GAS2
|
growth arrest-specific 2
|
|
chr3_-_56698073
|
0.104
|
NM_015224
|
FAM208A
|
family with sequence similarity 208, member A
|
|
chr3_+_142375738
|
0.102
|
NM_001172312
|
PLS1
|
plastin 1
|
|
chr17_+_67498391
|
0.102
|
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr20_+_32150139
|
0.101
|
NM_001039709
NM_005093
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr6_+_132891460
|
0.101
|
NM_175067
|
TAAR6
|
trace amine associated receptor 6
|
|
chr19_-_21950322
|
0.100
|
NM_173531
|
ZNF100
|
zinc finger protein 100
|
|
chr3_-_57678594
|
0.097
|
NM_152678
|
FAM116A
|
family with sequence similarity 116, member A
|
|
chr2_+_176969233
|
0.097
|
|
|
|
|
chr15_+_74422742
|
0.097
|
NM_020851
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr1_-_197036350
|
0.097
|
NM_001994
|
F13B
|
coagulation factor XIII, B polypeptide
|
|
chr3_+_186330859
|
0.096
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr17_-_66951381
|
0.094
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr6_-_150057659
|
0.093
|
|
NUP43
|
nucleoporin 43kDa
|
|
chr8_-_38205652
|
0.093
|
|
WHSC1L1
|
Wolf-Hirschhorn syndrome candidate 1-like 1
|
|
chr6_+_42788793
|
0.090
|
NM_015349
|
KIAA0240
|
KIAA0240
|
|
chr1_+_198608216
|
0.089
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr13_+_108921976
|
0.088
|
NM_001145645
NM_006573
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|
|
chr4_-_144826659
|
0.088
|
NM_002102
NM_198682
|
GYPE
|
glycophorin E (MNS blood group)
|
|
chr4_-_140060601
|
0.088
|
NM_201999
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chr15_-_31393906
|
0.086
|
NM_001252024
NM_001252030
NM_002420
|
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1
|
|
chr3_-_185826748
|
0.086
|
NM_004454
|
ETV5
|
ets variant 5
|
|
chr16_+_82068958
|
0.086
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chrX_+_47053213
|
0.085
|
|
UBA1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr17_+_3377346
|
0.084
|
NM_001128085
|
ASPA
|
aspartoacylase
|
|
chrX_+_47053192
|
0.083
|
NM_003334
|
UBA1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr16_-_31439650
|
0.083
|
NM_005205
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2
|
|
chr17_-_54893249
|
0.082
|
NM_001085430
|
C17orf67
|
chromosome 17 open reading frame 67
|
|
chr18_+_616671
|
0.082
|
NM_199167
|
CLUL1
|
clusterin-like 1 (retinal)
|
|
chrX_-_100548044
|
0.081
|
NM_024885
|
TAF7L
|
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa
|
|
chr8_-_12973752
|
0.081
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr12_+_20963637
|
0.081
|
NM_019844
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3
|
|
chr7_+_39017608
|
0.080
|
NM_001166018
NM_007252
|
POU6F2
|
POU class 6 homeobox 2
|
|
chr12_-_7848324
|
0.080
|
NM_020634
|
GDF3
|
growth differentiation factor 3
|
|
chr10_+_29577989
|
0.079
|
NM_032517
|
LYZL1
|
lysozyme-like 1
|
|
chr16_+_82068910
|
0.077
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chrX_-_32173585
|
0.077
|
NM_004013
NM_004020
NM_004021
NM_004022
NM_004023
|
DMD
|
dystrophin
|
|
chr1_-_160617042
|
0.077
|
NM_003037
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1
|
|
chr2_-_106055229
|
0.077
|
NM_201557
|
FHL2
|
four and a half LIM domains 2
|
|
chr7_-_38948776
|
0.077
|
NM_014396
NM_080631
|
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae)
|
|
chrX_+_150884506
|
0.076
|
NM_033085
|
FATE1
|
fetal and adult testis expressed 1
|
|
chr7_-_99766191
|
0.075
|
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4
|
|
chr7_-_100425035
|
0.075
|
NM_004444
|
EPHB4
|
EPH receptor B4
|
|
chr6_-_49834188
|
0.073
|
NM_001131
NM_170609
|
CRISP1
|
cysteine-rich secretory protein 1
|
|
chr21_-_31744546
|
0.073
|
NM_181621
|
KRTAP13-2
|
keratin associated protein 13-2
|
|
chr6_+_154407641
|
0.071
|
NM_001145287
|
OPRM1
|
opioid receptor, mu 1
|
|
chr17_-_10372875
|
0.070
|
NM_017533
|
MYH4
|
myosin, heavy chain 4, skeletal muscle
|
|
chr4_-_72649734
|
0.070
|
NM_000583
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chrX_-_33357725
|
0.070
|
NM_000109
|
DMD
|
dystrophin
|
|
chrX_+_47053227
|
0.068
|
|
UBA1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr11_+_87032428
|
0.067
|
|
TMEM135
|
transmembrane protein 135
|
|
chr7_-_64023444
|
0.067
|
|
|
|
|
chrX_+_47053271
|
0.067
|
|
UBA1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr3_+_142442855
|
0.066
|
|
|
|
|
chr10_+_5005601
|
0.066
|
|
AKR1C1
AKR1C3
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr3_-_105421044
|
0.065
|
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr6_-_29323971
|
0.065
|
NM_030876
|
OR5V1
|
olfactory receptor, family 5, subfamily V, member 1
|
|
chr7_-_99766238
|
0.065
|
NM_024637
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4
|
|
chr5_+_161274196
|
0.065
|
NM_000806
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr3_+_186330711
|
0.065
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr5_+_140220811
|
0.064
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr7_+_94292737
|
0.064
|
|
PEG10
|
paternally expressed 10
|
|
chr7_-_41742666
|
0.063
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr10_-_11574273
|
0.063
|
NM_001080491
|
USP6NL
|
USP6 N-terminal like
|
|
chr1_+_186798031
|
0.063
|
NM_024420
|
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr2_+_201756639
|
0.062
|
NM_001142356
|
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae)
|
|
chr17_-_29641068
|
0.062
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr10_-_128209996
|
0.061
|
NM_001004298
|
C10orf90
|
chromosome 10 open reading frame 90
|
|
chr6_-_138189313
|
0.061
|
|
|
|
|
chr4_-_2230957
|
0.061
|
NM_181808
|
POLN
|
polymerase (DNA directed) nu
|
|
chr2_+_28718937
|
0.060
|
NM_001170585
NM_153021
|
PLB1
|
phospholipase B1
|
|
chr3_+_180320955
|
0.060
|
|
TTC14
|
tetratricopeptide repeat domain 14
|
|
chr2_-_183387063
|
0.060
|
NM_005019
NM_001003683
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr16_-_71323090
|
0.059
|
NM_001099642
NM_018348
|
FTSJD1
|
FtsJ methyltransferase domain containing 1
|
|
chr19_-_38188344
|
0.058
|
|
ZNF607
|
zinc finger protein 607
|
|
chr17_-_2996289
|
0.056
|
NM_002548
|
OR1D2
|
olfactory receptor, family 1, subfamily D, member 2
|
|
chrX_+_86772714
|
0.056
|
NM_019117
NM_057162
|
KLHL4
|
kelch-like 4 (Drosophila)
|
|
chr4_+_40198526
|
0.055
|
NM_004310
|
RHOH
|
ras homolog gene family, member H
|
|
chr1_-_160549271
|
0.054
|
NM_001184879
NM_001184881
NM_001184882
NM_003874
|
CD84
|
CD84 molecule
|
|
chr2_-_142889269
|
0.054
|
NM_018557
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr2_+_29038862
|
0.054
|
NM_001008779
|
SPDYA
|
speedy homolog A (Xenopus laevis)
|
|
chr10_+_74870209
|
0.053
|
NM_015901
|
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13
|
|
chr2_-_241080072
|
0.053
|
NM_148961
|
OTOS
|
otospiralin
|
|
chr1_+_167189948
|
0.053
|
NM_001198786
NM_002697
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr17_+_17608163
|
0.053
|
|
|
|
|
chr12_-_90049818
|
0.052
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr12_-_11175169
|
0.052
|
NM_176888
|
TAS2R19
|
taste receptor, type 2, member 19
|
|
chr9_-_99145876
|
0.051
|
|
SLC35D2
|
solute carrier family 35, member D2
|
|
chr12_-_59314069
|
0.050
|
NM_153377
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr2_+_202098165
|
0.050
|
NM_001080124
NM_001228
NM_033355
NM_033358
|
CASP8
|
caspase 8, apoptosis-related cysteine peptidase
|
|
chr7_+_120702818
|
0.049
|
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr5_+_92918924
|
0.049
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr14_+_32047121
|
0.048
|
NM_001201573
|
NUBPL
|
nucleotide binding protein-like
|
|
chr1_+_67773046
|
0.048
|
NM_001559
|
IL12RB2
|
interleukin 12 receptor, beta 2
|
|
chr7_+_107110501
|
0.047
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chr1_+_109255555
|
0.047
|
NM_001144937
|
FNDC7
|
fibronectin type III domain containing 7
|
|
chr10_+_32856650
|
0.047
|
NM_024688
|
C10orf68
|
chromosome 10 open reading frame 68
|
|
chr10_-_30918625
|
0.046
|
NM_183058
|
LYZL2
|
lysozyme-like 2
|
|
chr3_-_111314181
|
0.046
|
NM_024508
|
ZBED2
|
zinc finger, BED-type containing 2
|
|
chr20_-_20036685
|
0.046
|
NM_016652
|
CRNKL1
|
crooked neck pre-mRNA splicing factor-like 1 (Drosophila)
|
|
chr5_+_161274684
|
0.046
|
NM_001127643
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr1_+_18081807
|
0.046
|
NM_030812
|
ACTL8
|
actin-like 8
|