|
chr6_+_37137882
|
3.681
|
NM_001243186
NM_002648
|
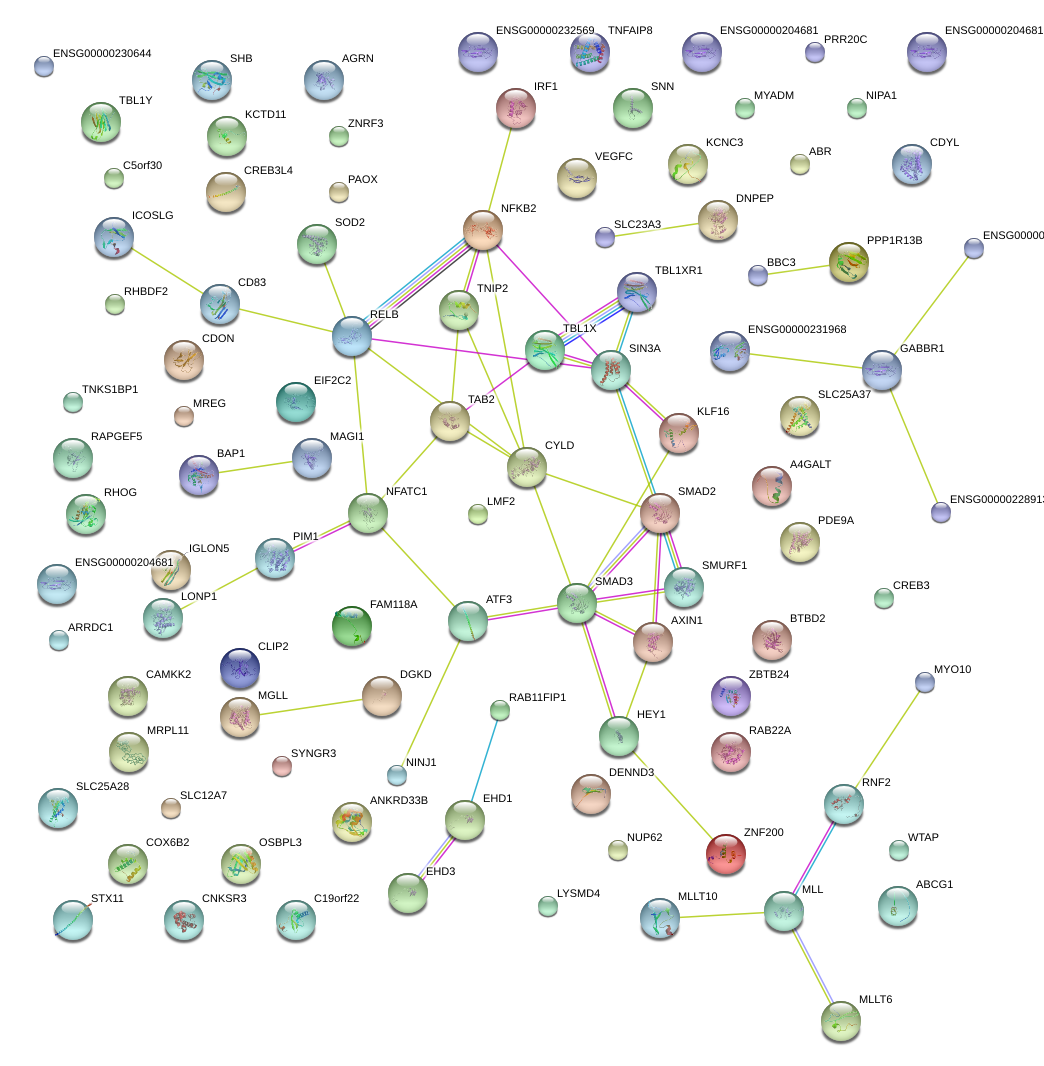
PIM1
|
pim-1 oncogene
|
|
chr2_-_220252569
|
3.545
|
|
DNPEP
|
aspartyl aminopeptidase
|
|
chr16_+_11762235
|
3.246
|
NM_003498
|
SNN
|
stannin
|
|
chr2_-_220252627
|
2.942
|
NM_012100
|
DNPEP
|
aspartyl aminopeptidase
|
|
chr6_+_4776826
|
2.854
|
|
CDYL
|
chromodomain protein, Y-like
|
|
chr10_+_104155431
|
2.760
|
NM_002502
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr21_+_44073650
|
2.635
|
NM_001001567
NM_001001568
NM_001001569
NM_001001570
NM_001001571
NM_001001572
NM_001001573
NM_001001574
NM_001001575
NM_001001576
NM_001001577
NM_001001578
NM_001001579
NM_001001580
NM_001001581
NM_001001582
NM_001001583
NM_001001584
NM_001001585
NM_002606
|
PDE9A
|
phosphodiesterase 9A
|
|
chr19_+_45504686
|
2.529
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr9_-_38069082
|
2.461
|
|
SHB
|
Src homology 2 domain containing adaptor protein B
|
|
chr5_+_118604417
|
2.408
|
NM_001077654
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8
|
|
chr21_+_43639202
|
2.267
|
NM_004915
NM_016818
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr6_+_4776641
|
2.242
|
NM_004824
|
CDYL
|
chromodomain protein, Y-like
|
|
chr7_+_73703704
|
2.227
|
NM_003388
NM_032421
|
CLIP2
|
CAP-GLY domain containing linker protein 2
|
|
chr2_-_220252513
|
2.217
|
|
DNPEP
|
aspartyl aminopeptidase
|
|
chr4_-_177713664
|
2.124
|
NM_005429
|
VEGFC
|
vascular endothelial growth factor C
|
|
chr15_+_67358161
|
1.981
|
|
SMAD3
|
SMAD family member 3
|
|
chr6_+_14118002
|
1.906
|
|
CD83
|
CD83 molecule
|
|
chr11_-_125933186
|
1.881
|
NM_001243597
NM_016952
|
CDON
|
Cdon homolog (mouse)
|
|
chr10_-_101380160
|
1.854
|
|
SLC25A28
|
solute carrier family 25, member 28
|
|
chr1_+_955609
|
1.841
|
|
AGRN
|
agrin
|
|
chr5_+_118604504
|
1.837
|
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8
|
|
chr4_-_2758018
|
1.796
|
NM_024309
|
TNIP2
|
TNFAIP3 interacting protein 2
|
|
chr8_-_80679897
|
1.769
|
NM_001040708
NM_012258
|
HEY1
|
hairy/enhancer-of-split related with YRPW motif 1
|
|
chr9_-_95896498
|
1.751
|
NM_004148
|
NINJ1
|
ninjurin 1
|
|
chr19_-_47734215
|
1.721
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr16_+_50776012
|
1.716
|
NM_001042412
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr6_-_29595746
|
1.684
|
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr17_-_1083008
|
1.683
|
NM_021962
|
ABR
|
active BCR-related gene
|
|
chr16_+_50775960
|
1.676
|
NM_001042355
NM_015247
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr5_-_16936290
|
1.662
|
|
MYO10
|
myosin X
|
|
chr15_+_67358194
|
1.644
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr1_+_955468
|
1.640
|
NM_198576
|
AGRN
|
agrin
|
|
chr6_-_109804322
|
1.629
|
|
ZBTB24
|
zinc finger and BTB domain containing 24
|
|
chr10_-_101380217
|
1.607
|
NM_031212
|
SLC25A28
|
solute carrier family 25, member 28
|
|
chr1_+_2518071
|
1.594
|
NM_001195736
NM_001195737
NM_001195738
NM_001195740
NM_001195741
NM_152371
|
FAM213B
|
family with sequence similarity 213, member B
|
|
chr9_+_140500148
|
1.582
|
|
ARRDC1
|
arrestin domain containing 1
|
|
chr11_-_64645925
|
1.582
|
|
EHD1
|
EH-domain containing 1
|
|
chr22_+_29279573
|
1.556
|
NM_001206998
|
ZNRF3
|
zinc and ring finger 3
|
|
chr22_-_43116796
|
1.526
|
NM_017436
|
A4GALT
|
alpha 1,4-galactosyltransferase
|
|
chrX_+_9432980
|
1.492
|
NM_005647
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr8_+_142138684
|
1.490
|
NM_014957
|
DENND3
|
DENN/MADD domain containing 3
|
|
chr10_+_135192694
|
1.470
|
NM_152911
NM_207127
NM_207128
|
PAOX
|
polyamine oxidase (exo-N4-amino)
|
|
chr20_+_56884839
|
1.465
|
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chr20_+_56885027
|
1.464
|
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chr16_+_2039945
|
1.451
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr7_-_25019625
|
1.446
|
NM_015550
NM_145320
NM_145321
NM_145322
|
OSBPL3
|
oxysterol binding protein-like 3
|
|
chr2_+_234263141
|
1.409
|
NM_152879
|
DGKD
|
diacylglycerol kinase, delta 130kDa
|
|
chr9_+_140500136
|
1.377
|
|
ARRDC1
|
arrestin domain containing 1
|
|
chr5_-_131826426
|
1.367
|
NM_002198
|
IRF1
|
interferon regulatory factor 1
|
|
chr1_+_212781969
|
1.365
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr14_-_104313730
|
1.359
|
NM_015316
|
PPP1R13B
|
protein phosphatase 1, regulatory subunit 13B
|
|
chr8_-_37756971
|
1.357
|
NM_001002814
NM_025151
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chr3_-_127541160
|
1.350
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr3_-_45267804
|
1.309
|
NM_015444
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr1_+_2518464
|
1.307
|
|
FAM213B
|
family with sequence similarity 213, member B
|
|
chr5_+_10564431
|
1.304
|
NM_001164440
|
ANKRD33B
|
ankyrin repeat domain 33B
|
|
chr11_-_3862137
|
1.289
|
|
RHOG
|
ras homolog gene family, member G (rho G)
|
|
chr15_-_23086290
|
1.272
|
NM_144599
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr21_-_45660841
|
1.269
|
|
ICOSLG
|
inducible T-cell co-stimulator ligand
|
|
chr15_-_100273540
|
1.261
|
|
LYSMD4
|
LysM, putative peptidoglycan-binding, domain containing 4
|
|
chr19_-_50432952
|
1.259
|
NM_016553
|
NUP62
|
nucleoporin 62kDa
|
|
chr10_-_101380086
|
1.257
|
|
SLC25A28
|
solute carrier family 25, member 28
|
|
chr22_-_50946020
|
1.256
|
|
LMF2
|
lipase maturation factor 2
|
|
chr12_-_121734543
|
1.255
|
NM_172226
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr16_-_402617
|
1.239
|
NM_003502
NM_181050
|
AXIN1
|
axin 1
|
|
chr19_+_54371150
|
1.235
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr18_+_77155927
|
1.233
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr7_-_98741375
|
1.229
|
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1
|
|
chr16_-_3285359
|
1.229
|
NM_001145448
NM_198088
NM_001145446
NM_198087
|
ZNF200
|
zinc finger protein 200
|
|
chr9_+_35732207
|
1.227
|
NM_006368
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr17_-_74497406
|
1.219
|
NM_024599
|
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila)
|
|
chr18_+_77155966
|
1.211
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr6_+_149639054
|
1.209
|
|
TAB2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2
|
|
chr10_+_74451838
|
1.201
|
NM_138357
|
MCU
|
mitochondrial calcium uniporter
|
|
chr5_+_102594406
|
1.199
|
NM_033211
|
C5orf30
|
chromosome 5 open reading frame 30
|
|
chr5_-_1112106
|
1.191
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr19_-_50832633
|
1.187
|
NM_004977
|
KCNC3
|
potassium voltage-gated channel, Shaw-related subfamily, member 3
|
|
chr11_-_575848
|
1.186
|
|
LOC143666
|
uncharacterized LOC143666
|
|
chr2_+_64681270
|
1.179
|
NM_014181
|
LGALSL
|
lectin, galactoside-binding-like
|
|
chr6_-_160114318
|
1.178
|
NM_000636
NM_001024465
NM_001024466
|
SOD2
|
superoxide dismutase 2, mitochondrial
|
|
chr7_-_22396511
|
1.178
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr11_-_66234176
|
1.175
|
|
MRPL11
|
mitochondrial ribosomal protein L11
|
|
chr9_+_140500054
|
1.171
|
NM_152285
|
ARRDC1
|
arrestin domain containing 1
|
|
chr21_-_45660699
|
1.168
|
NM_015259
|
ICOSLG
|
inducible T-cell co-stimulator ligand
|
|
chr17_+_36861857
|
1.167
|
NM_005937
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr6_+_144471631
|
1.155
|
NM_003764
|
STX11
|
syntaxin 11
|
|
chr11_-_57092283
|
1.151
|
NM_033396
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa
|
|
chr15_-_75743776
|
1.149
|
NM_015477
|
SIN3A
|
SIN3 transcription regulator homolog A (yeast)
|
|
chr17_+_7255427
|
1.148
|
|
KCTD11
|
potassium channel tetramerisation domain containing 11
|
|
chr8_-_141645603
|
1.130
|
NM_001164623
NM_012154
|
EIF2C2
|
eukaryotic translation initiation factor 2C, 2
|
|
chr9_+_140500097
|
1.130
|
|
ARRDC1
|
arrestin domain containing 1
|
|
chr22_+_45705965
|
1.130
|
|
FAM118A
|
family with sequence similarity 118, member A
|
|
chr11_+_118307072
|
1.129
|
NM_001197104
NM_005933
|
MLL
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)
|
|
chr11_-_64646053
|
1.121
|
NM_006795
|
EHD1
|
EH-domain containing 1
|
|
chr3_-_52443893
|
1.112
|
NM_004656
|
BAP1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase)
|
|
chr6_-_30654978
|
1.110
|
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr3_-_45267620
|
1.109
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr12_-_121734474
|
1.104
|
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr21_+_43639930
|
1.102
|
NM_207174
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr19_-_1863425
|
1.102
|
|
KLF16
|
Kruppel-like factor 16
|
|
chr8_+_23386362
|
1.100
|
NM_016612
|
SLC25A37
|
solute carrier family 25, member 37
|
|
chr6_+_160148608
|
1.099
|
NM_152857
|
WTAP
|
Wilms tumor 1 associated protein
|
|
chr19_+_51815095
|
1.090
|
NM_001101372
|
IGLON5
|
IgLON family member 5
|
|
chr19_-_2015628
|
1.088
|
NM_017797
|
BTBD2
|
BTB (POZ) domain containing 2
|
|
chr19_-_55866063
|
1.077
|
NM_144613
|
COX6B2
|
cytochrome c oxidase subunit VIb polypeptide 2 (testis)
|
|
chr3_+_122399665
|
1.073
|
NM_017554
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14
|
|
chr9_-_38069207
|
1.066
|
NM_003028
|
SHB
|
Src homology 2 domain containing adaptor protein B
|
|
chr3_-_52444016
|
1.066
|
|
BAP1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase)
|
|
chr17_-_17726855
|
1.058
|
|
SREBF1
|
sterol regulatory element binding transcription factor 1
|
|
chr9_+_100174179
|
1.043
|
NM_014290
|
TDRD7
|
tudor domain containing 7
|
|
chr7_-_1499107
|
1.041
|
NM_182924
|
MICALL2
|
MICAL-like 2
|
|
chr5_+_17217474
|
1.037
|
NM_006317
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr1_-_935469
|
1.030
|
NM_001142467
NM_021170
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr5_-_32313069
|
1.030
|
NM_001040446
|
MTMR12
|
myotubularin related protein 12
|
|
chr22_+_45705766
|
1.026
|
NM_017911
|
FAM118A
|
family with sequence similarity 118, member A
|
|
chr1_+_206858308
|
1.025
|
NM_004759
NM_032960
|
MAPKAPK2
|
mitogen-activated protein kinase-activated protein kinase 2
|
|
chr6_+_138188352
|
1.021
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr15_-_100273620
|
1.021
|
NM_152449
|
LYSMD4
|
LysM, putative peptidoglycan-binding, domain containing 4
|
|
chr5_-_32312943
|
1.019
|
|
MTMR12
|
myotubularin related protein 12
|
|
chr21_-_45759234
|
1.015
|
|
C21orf2
|
chromosome 21 open reading frame 2
|
|
chr6_+_14117857
|
1.013
|
NM_001040280
NM_004233
|
CD83
|
CD83 molecule
|
|
chr6_+_31126340
|
1.012
|
|
TCF19
|
transcription factor 19
|
|
chr6_-_13408368
|
1.010
|
NM_001242630
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr7_+_28448970
|
1.009
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr19_+_50879681
|
1.007
|
NM_007121
|
NR1H2
|
nuclear receptor subfamily 1, group H, member 2
|
|
chr20_+_61427804
|
1.004
|
NM_018270
|
C20orf20
|
chromosome 20 open reading frame 20
|
|
chr1_+_212782103
|
1.004
|
|
ATF3
|
activating transcription factor 3
|
|
chr6_+_31126114
|
1.002
|
NM_001077511
NM_007109
|
TCF19
|
transcription factor 19
|
|
chr15_-_68521921
|
1.000
|
NM_017882
|
CLN6
|
ceroid-lipofuscinosis, neuronal 6, late infantile, variant
|
|
chr14_+_21538339
|
0.996
|
NM_018071
|
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40
|
|
chr2_+_234263153
|
0.993
|
|
DGKD
|
diacylglycerol kinase, delta 130kDa
|
|
chr11_+_65292576
|
0.991
|
|
SCYL1
|
SCY1-like 1 (S. cerevisiae)
|
|
chr7_-_100487273
|
0.991
|
NM_001015072
|
UFSP1
|
UFM1-specific peptidase 1 (non-functional)
|
|
chr1_-_156721280
|
0.988
|
|
HDGF
|
hepatoma-derived growth factor
|
|
chr22_-_50946129
|
0.986
|
NM_033200
|
LMF2
|
lipase maturation factor 2
|
|
chr2_+_64681610
|
0.978
|
|
LGALSL
|
lectin, galactoside-binding-like
|
|
chr10_-_135171512
|
0.976
|
NM_001098483
NM_198472
|
C10orf125
|
chromosome 10 open reading frame 125
|
|
chr8_+_104383701
|
0.972
|
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr6_-_109804432
|
0.970
|
NM_001164313
NM_014797
|
ZBTB24
|
zinc finger and BTB domain containing 24
|
|
chr17_-_48207113
|
0.969
|
NM_174920
|
SAMD14
|
sterile alpha motif domain containing 14
|
|
chr20_+_30946786
|
0.968
|
|
ASXL1
|
additional sex combs like 1 (Drosophila)
|
|
chr17_+_39382878
|
0.967
|
NM_031961
|
KRTAP9-2
|
keratin associated protein 9-2
|
|
chr6_+_37787592
|
0.967
|
|
ZFAND3
|
zinc finger, AN1-type domain 3
|
|
chr6_+_18155535
|
0.962
|
NM_153042
|
KDM1B
|
lysine (K)-specific demethylase 1B
|
|
chr19_+_6739697
|
0.962
|
NM_004240
|
TRIP10
|
thyroid hormone receptor interactor 10
|
|
chr1_-_160068359
|
0.961
|
NM_052868
|
IGSF8
|
immunoglobulin superfamily, member 8
|
|
chr6_-_160114186
|
0.948
|
|
SOD2
|
superoxide dismutase 2, mitochondrial
|
|
chr5_+_179234002
|
0.947
|
NM_001142299
|
SQSTM1
|
sequestosome 1
|
|
chr8_+_145064583
|
0.938
|
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding)
|
|
chr6_-_44233244
|
0.933
|
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon
|
|
chr22_-_45636760
|
0.933
|
|
KIAA0930
|
KIAA0930
|
|
chr2_-_288181
|
0.928
|
NM_001002919
|
FAM150B
|
family with sequence similarity 150, member B
|
|
chr11_-_72353435
|
0.928
|
NM_001143839
NM_001243784
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr9_-_132805256
|
0.927
|
|
FNBP1
|
formin binding protein 1
|
|
chr12_-_133306531
|
0.920
|
|
ANKLE2
|
ankyrin repeat and LEM domain containing 2
|
|
chr20_+_35974458
|
0.920
|
NM_198291
|
SRC
|
v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian)
|
|
chr17_-_53499055
|
0.919
|
NM_012329
|
MMD
|
monocyte to macrophage differentiation-associated
|
|
chr19_+_54372659
|
0.919
|
NM_001020821
NM_001020818
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr6_-_30655078
|
0.912
|
NM_001134870
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr7_+_55086922
|
0.910
|
|
EGFR
|
epidermal growth factor receptor
|
|
chr10_+_82214066
|
0.907
|
|
TSPAN14
|
tetraspanin 14
|
|
chr17_+_36861398
|
0.907
|
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr2_+_85360373
|
0.906
|
NM_031283
|
TCF7L1
|
transcription factor 7-like 1 (T-cell specific, HMG-box)
|
|
chr10_-_105614952
|
0.905
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr17_-_80606095
|
0.905
|
NM_019613
|
WDR45L
|
WDR45-like
|
|
chr14_-_21566728
|
0.900
|
NM_016423
|
ZNF219
|
zinc finger protein 219
|
|
chr1_+_166808721
|
0.899
|
NM_017542
|
POGK
|
pogo transposable element with KRAB domain
|
|
chr9_+_100174369
|
0.896
|
|
TDRD7
|
tudor domain containing 7
|
|
chr1_-_161102210
|
0.896
|
NM_001039712
|
DEDD
|
death effector domain containing
|
|
chr4_-_2757750
|
0.891
|
NM_001161527
|
TNIP2
|
TNFAIP3 interacting protein 2
|
|
chr6_-_13486986
|
0.891
|
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr14_-_21566497
|
0.890
|
|
ZNF219
|
zinc finger protein 219
|
|
chr17_+_7255207
|
0.884
|
NM_001002914
|
KCTD11
|
potassium channel tetramerisation domain containing 11
|
|
chr1_-_161102367
|
0.883
|
|
DEDD
|
death effector domain containing
|
|
chr10_+_82213892
|
0.880
|
NM_001128309
NM_030927
|
TSPAN14
|
tetraspanin 14
|
|
chr8_-_494834
|
0.880
|
|
C8orf42
|
chromosome 8 open reading frame 42
|
|
chr1_+_100111430
|
0.877
|
NM_017734
|
PALMD
|
palmdelphin
|
|
chr4_-_37687932
|
0.874
|
NM_001085399
NM_001085400
|
RELL1
|
RELT-like 1
|
|
chr12_-_50101192
|
0.873
|
NM_175736
NM_198900
|
FMNL3
|
formin-like 3
|
|
chr19_+_41725277
|
0.873
|
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr17_-_80231240
|
0.869
|
|
CSNK1D
|
casein kinase 1, delta
|
|
chr19_-_55658310
|
0.869
|
|
TNNT1
|
troponin T type 1 (skeletal, slow)
|
|
chr17_-_73874634
|
0.861
|
NM_033452
|
TRIM47
|
tripartite motif containing 47
|
|
chr19_-_47922339
|
0.859
|
|
MEIS3
|
Meis homeobox 3
|
|
chr1_-_160068617
|
0.857
|
NM_001206665
|
IGSF8
|
immunoglobulin superfamily, member 8
|
|
chr19_+_47778099
|
0.851
|
NM_178511
|
PRR24
|
proline rich 24
|
|
chr12_-_124068864
|
0.846
|
|
|
|
|
chr11_+_369723
|
0.845
|
NM_178537
|
B4GALNT4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4
|
|
chr6_+_14117486
|
0.844
|
NM_001251901
|
CD83
|
CD83 molecule
|
|
chr19_-_10444186
|
0.843
|
NM_133452
|
RAVER1
|
ribonucleoprotein, PTB-binding 1
|
|
chr20_-_62462568
|
0.841
|
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr22_+_38302296
|
0.841
|
|
MICALL1
|
MICAL-like 1
|
|
chr20_-_61569227
|
0.839
|
|
DIDO1
|
death inducer-obliterator 1
|
|
chr1_-_229761716
|
0.837
|
|
TAF5L
|
TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa
|
|
chr16_-_402398
|
0.836
|
|
AXIN1
|
axin 1
|
|
chr19_+_10197023
|
0.836
|
|
C19orf66
|
chromosome 19 open reading frame 66
|
|
chr1_-_36851484
|
0.835
|
NM_032017
|
STK40
|
serine/threonine kinase 40
|
|
chr3_-_64673330
|
0.834
|
NM_182920
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9
|
|
chr4_+_38665587
|
0.831
|
NM_016531
|
KLF3
|
Kruppel-like factor 3 (basic)
|
|
chr7_+_45039777
|
0.830
|
NM_001167934
NM_001167935
NM_031443
|
CCM2
|
cerebral cavernous malformation 2
|