|
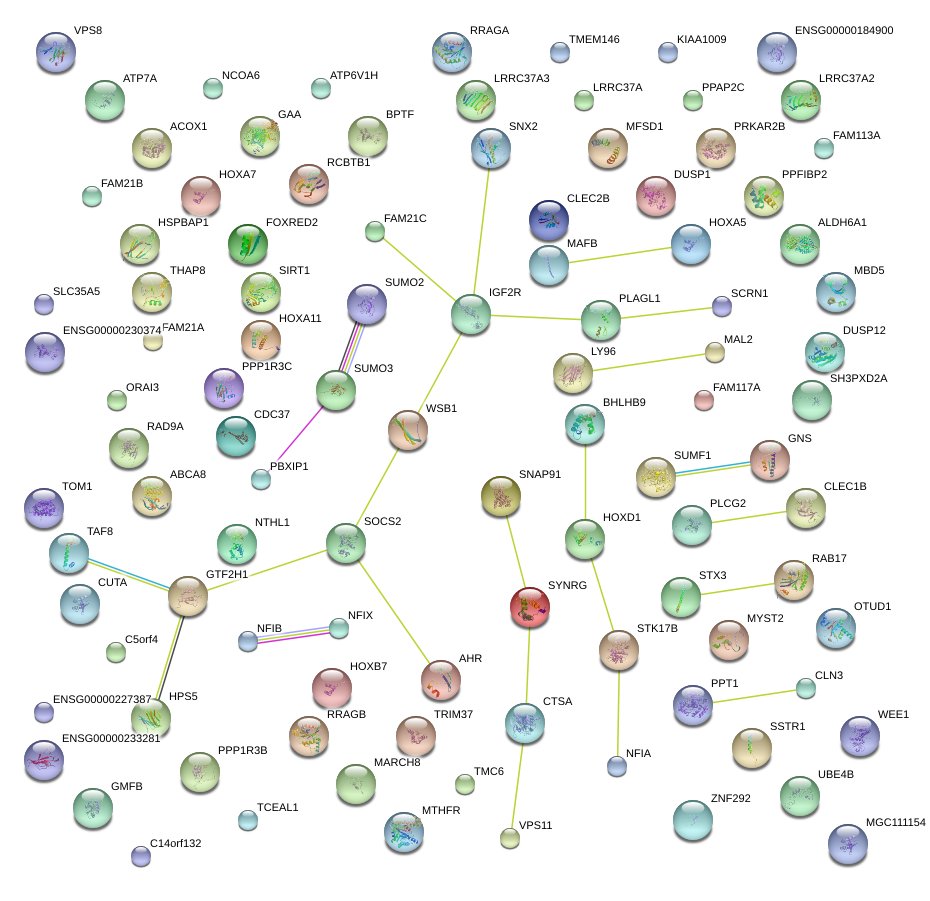
chr17_-_46688362
|
12.788
|
NM_004502
|
HOXB7
|
homeobox B7
|
|
chr17_-_46688279
|
12.009
|
|
HOXB7
|
homeobox B7
|
|
chr20_-_39317875
|
11.060
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr1_-_11866079
|
8.419
|
NM_005957
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chrX_+_101975641
|
7.465
|
NM_001142524
NM_001142525
NM_001142526
NM_001142527
NM_001142528
NM_030639
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9
|
|
chr17_-_46687925
|
7.456
|
|
HOXB7
|
homeobox B7
|
|
chr12_+_93964801
|
7.448
|
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr3_-_122512526
|
7.386
|
NM_024610
|
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1
|
|
chr17_+_25621105
|
6.572
|
NM_015626
NM_134265
|
WSB1
|
WD repeat and SOCS box containing 1
|
|
chr4_+_100871625
|
6.564
|
|
LOC256880
|
uncharacterized LOC256880
|
|
chr1_+_61869699
|
6.525
|
|
NFIA
|
nuclear factor I/A
|
|
chr5_-_172198160
|
6.240
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chrX_+_55744049
|
6.183
|
NM_006064
NM_016656
|
RRAGB
|
Ras-related GTP binding B
|
|
chr6_+_160390128
|
6.065
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chrX_+_77166178
|
6.049
|
NM_000052
|
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide
|
|
chr12_-_10021932
|
5.964
|
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr17_+_65821625
|
5.927
|
NM_004459
NM_182641
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr1_-_154928564
|
5.875
|
NM_020524
|
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1
|
|
chr11_+_67159421
|
5.786
|
NM_004584
|
RAD9A
|
RAD9 homolog A (S. pombe)
|
|
chr3_+_112280871
|
5.677
|
NM_017945
|
SLC35A5
|
solute carrier family 35, member A5
|
|
chr6_-_33386064
|
5.646
|
NM_001014433
NM_001014837
NM_001014838
NM_001014840
NM_015921
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr6_-_84418705
|
5.586
|
NM_001242792
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr7_-_27183225
|
5.484
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr8_+_120220554
|
5.388
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr11_-_18343670
|
5.194
|
NM_007216
NM_181507
NM_181508
|
HPS5
|
Hermansky-Pudlak syndrome 5
|
|
chr10_-_93392857
|
5.085
|
NM_005398
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr6_+_87865261
|
4.992
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr6_-_84419108
|
4.911
|
NM_001242793
NM_001242794
NM_014841
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr8_-_9009050
|
4.887
|
NM_001201329
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B
|
|
chr6_-_84937313
|
4.885
|
NM_014895
|
KIAA1009
|
KIAA1009
|
|
chr16_+_5083671
|
4.874
|
|
LOC100507589
|
uncharacterized LOC100507589
|
|
chr12_-_65153182
|
4.819
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr6_-_84419018
|
4.802
|
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr22_-_36903086
|
4.793
|
NM_024955
NM_001102371
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2
|
|
chr11_+_59522801
|
4.581
|
|
STX3
|
syntaxin 3
|
|
chr17_+_78075299
|
4.543
|
NM_000152
NM_001079803
NM_001079804
|
GAA
|
glucosidase, alpha; acid
|
|
chr2_-_238499317
|
4.496
|
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr14_-_54955664
|
4.484
|
|
GMFB
|
glia maturation factor, beta
|
|
chr17_-_47841517
|
4.483
|
NM_030802
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr17_-_62915580
|
4.423
|
|
LRRC37A3
|
leucine rich repeat containing 37, member A3
|
|
chr17_-_47841468
|
4.408
|
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr11_+_9595451
|
4.389
|
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr6_+_42018264
|
4.367
|
|
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa
|
|
chrX_+_102883891
|
4.316
|
NM_001006639
NM_004780
|
TCEAL1
|
transcription elongation factor A (SII)-like 1
|
|
chr13_-_50159628
|
4.309
|
|
RCBTB1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1
|
|
chr11_+_59522905
|
4.291
|
|
STX3
|
syntaxin 3
|
|
chr14_+_38677186
|
4.232
|
NM_001049
|
SSTR1
|
somatostatin receptor 1
|
|
chr5_+_122110778
|
4.221
|
|
SNX2
|
sorting nexin 2
|
|
chr17_+_78075390
|
4.173
|
|
GAA
|
glucosidase, alpha; acid
|
|
chr5_+_122110762
|
4.168
|
|
SNX2
|
sorting nexin 2
|
|
chr6_-_33385814
|
4.134
|
|
|
|
|
chr7_+_106685007
|
4.130
|
NM_002736
|
PRKAR2B
|
protein kinase, cAMP-dependent, regulatory, type II, beta
|
|
chr14_-_54955714
|
4.026
|
NM_004124
|
GMFB
|
glia maturation factor, beta
|
|
chr10_-_46090191
|
3.951
|
NM_001002265
NM_145021
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8
|
|
chr11_+_7535213
|
3.936
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr20_+_44520021
|
3.880
|
|
CTSA
|
cathepsin A
|
|
chr5_-_154230157
|
3.847
|
|
C5orf4
|
chromosome 5 open reading frame 4
|
|
chr3_+_112280911
|
3.830
|
|
SLC35A5
|
solute carrier family 35, member A5
|
|
chr17_-_76124767
|
3.803
|
|
TMC6
|
transmembrane channel-like 6
|
|
chr8_-_54755821
|
3.759
|
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr8_-_54755546
|
3.747
|
NM_213620
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr20_-_2821245
|
3.743
|
NM_022760
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr12_-_65153148
|
3.733
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr11_+_7535000
|
3.699
|
NM_003621
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr8_-_54755836
|
3.694
|
NM_015941
NM_213619
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr7_-_27196163
|
3.649
|
NM_006896
|
HOXA7
|
homeobox A7
|
|
chr5_-_154230212
|
3.605
|
NM_032385
|
C5orf4
|
chromosome 5 open reading frame 4
|
|
chr17_-_57184116
|
3.580
|
|
TRIM37
|
tripartite motif containing 37
|
|
chr10_+_69644341
|
3.572
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chr17_+_25621141
|
3.563
|
|
WSB1
|
WD repeat and SOCS box containing 1
|
|
chr8_+_74903563
|
3.557
|
NM_001195797
NM_015364
|
LY96
|
lymphocyte antigen 96
|
|
chr17_-_35969401
|
3.540
|
NM_001163544
NM_001163545
NM_001163546
NM_001163547
NM_007247
NM_080550
NM_198882
|
SYNRG
|
synergin, gamma
|
|
chr19_-_36545298
|
3.531
|
|
THAP8
|
THAP domain containing 8
|
|
chr7_-_27224762
|
3.529
|
NM_005523
|
HOXA11
|
homeobox A11
|
|
chr12_-_65153055
|
3.527
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr1_-_154928595
|
3.514
|
|
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1
|
|
chrX_+_77166223
|
3.498
|
|
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide
|
|
chr2_+_148778573
|
3.490
|
NM_018328
|
MBD5
|
methyl-CpG binding domain protein 5
|
|
chr5_+_122110737
|
3.489
|
NM_003100
|
SNX2
|
sorting nexin 2
|
|
chr17_-_73975507
|
3.451
|
NM_001185039
NM_004035
NM_007292
|
ACOX1
|
acyl-CoA oxidase 1, palmitoyl
|
|
chr3_-_4508939
|
3.450
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr11_+_9595429
|
3.450
|
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr3_+_158519888
|
3.420
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr3_-_4508928
|
3.362
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr6_+_42018250
|
3.337
|
NM_138572
|
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa
|
|
chr10_-_105452878
|
3.312
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr14_-_74551159
|
3.277
|
NM_005589
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr16_-_28503079
|
3.259
|
NM_000086
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3
|
|
chr22_+_35695872
|
3.254
|
|
TOM1
|
target of myb1 (chicken)
|
|
chr19_-_291168
|
3.236
|
NM_177543
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr1_+_10092990
|
3.209
|
NM_001105562
NM_006048
|
UBE4B
|
ubiquitination factor E4B
|
|
chr16_+_81812897
|
3.191
|
NM_002661
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr3_-_4508955
|
3.172
|
NM_001164674
NM_001164675
NM_182760
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr12_-_65153189
|
3.172
|
NM_002076
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr3_+_184529871
|
3.120
|
NM_001009921
NM_015303
|
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae)
|
|
chr11_+_9595183
|
3.109
|
NM_003390
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr16_+_30960413
|
3.090
|
|
ORAI3
|
ORAI calcium release-activated calcium modulator 3
|
|
chr21_-_46237978
|
3.089
|
|
SUMO3
|
SMT3 suppressor of mif two 3 homolog 3 (S. cerevisiae)
|
|
chr10_+_23728197
|
3.075
|
NM_001145373
|
OTUD1
|
OTU domain containing 1
|
|
chr16_+_30960377
|
3.066
|
NM_152288
|
ORAI3
|
ORAI calcium release-activated calcium modulator 3
|
|
chr16_+_30960434
|
3.057
|
|
ORAI3
|
ORAI calcium release-activated calcium modulator 3
|
|
chr16_-_28503622
|
3.051
|
NM_001042432
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3
|
|
chr7_-_30029284
|
3.047
|
NM_001145515
NM_014766
NM_001145513
|
SCRN1
|
secernin 1
|
|
chr16_-_2097818
|
3.046
|
NM_002528
|
NTHL1
|
nth endonuclease III-like 1 (E. coli)
|
|
chr7_+_27225009
|
3.036
|
|
HOXA11-AS1
|
HOXA11 antisense RNA 1 (non-protein coding)
|
|
chr11_+_118938530
|
3.006
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr2_+_177053306
|
2.966
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr1_-_40562909
|
2.965
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr17_+_47865980
|
2.962
|
NM_001199155
NM_001199156
NM_001199157
NM_001199158
NM_007067
|
KAT7
|
K(lysine) acetyltransferase 7
|
|
chr11_+_118938528
|
2.935
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr16_+_81812899
|
2.934
|
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr1_-_40562917
|
2.932
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr11_+_9595309
|
2.926
|
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr19_+_5720687
|
2.913
|
NM_152784
|
TMEM146
|
transmembrane protein 146
|
|
chr17_-_66951381
|
2.906
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr10_+_51827710
|
2.885
|
|
FAM21A
|
family with sequence similarity 21, member A
|
|
chr20_-_2821120
|
2.883
|
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr2_-_197036301
|
2.872
|
NM_004226
|
STK17B
|
serine/threonine kinase 17b
|
|
chr7_+_17338182
|
2.859
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr3_+_158519911
|
2.855
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr10_+_69644708
|
2.850
|
|
SIRT1
|
sirtuin 1
|
|
chr17_-_73975443
|
2.843
|
|
ACOX1
|
acyl-CoA oxidase 1, palmitoyl
|
|
chr20_+_44519590
|
2.839
|
NM_000308
NM_001127695
NM_001167594
|
CTSA
|
cathepsin A
|
|
chr17_+_47866061
|
2.828
|
|
KAT7
|
K(lysine) acetyltransferase 7
|
|
chr14_+_96505566
|
2.823
|
NM_001252507
|
C14orf132
|
chromosome 14 open reading frame 132
|
|
chr1_-_11865963
|
2.821
|
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr16_+_81812925
|
2.792
|
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr11_+_118938515
|
2.757
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr6_-_144329342
|
2.716
|
NM_001080952
NM_001080953
NM_001080954
NM_001080956
NM_002656
NM_006718
|
PLAGL1
HYMAI
|
pleiomorphic adenoma gene-like 1
hydatidiform mole associated and imprinted (non-protein coding)
|
|
chr11_+_18343794
|
2.710
|
NM_001142307
NM_005316
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr5_+_172483285
|
2.709
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr9_-_126030854
|
2.688
|
NM_001171137
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr16_-_23521755
|
2.657
|
|
GGA2
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 2
|
|
chr19_-_36545119
|
2.623
|
|
THAP8
|
THAP domain containing 8
|
|
chr6_+_123110345
|
2.620
|
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A
|
|
chr4_-_149363498
|
2.613
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr10_-_116418056
|
2.600
|
NM_002313
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr6_-_151773168
|
2.599
|
NM_017909
|
RMND1
|
required for meiotic nuclear division 1 homolog (S. cerevisiae)
|
|
chr9_-_126030792
|
2.596
|
NM_018387
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chrX_+_102883647
|
2.585
|
NM_001006640
|
TCEAL1
|
transcription elongation factor A (SII)-like 1
|
|
chrX_+_118108575
|
2.582
|
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr4_-_76439582
|
2.580
|
NM_001008925
NM_001009922
NM_015436
|
RCHY1
|
ring finger and CHY zinc finger domain containing 1
|
|
chr12_+_12869818
|
2.577
|
NM_004064
|
CDKN1B
|
cyclin-dependent kinase inhibitor 1B (p27, Kip1)
|
|
chr8_-_95274540
|
2.556
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr16_+_2570322
|
2.546
|
NM_001145815
NM_015944
|
AMDHD2
|
amidohydrolase domain containing 2
|
|
chr16_-_4588379
|
2.540
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr1_+_10093149
|
2.536
|
|
UBE4B
|
ubiquitination factor E4B
|
|
chr20_-_36156226
|
2.535
|
|
BLCAP
|
bladder cancer associated protein
|
|
chr7_-_100808832
|
2.512
|
NM_003378
|
VGF
|
VGF nerve growth factor inducible
|
|
chrX_-_100872926
|
2.508
|
NM_001009584
NM_001184768
NM_019007
|
ARMCX6
|
armadillo repeat containing, X-linked 6
|
|
chr5_+_68710938
|
2.508
|
NM_001038603
NM_001244734
|
MARVELD2
|
MARVEL domain containing 2
|
|
chr3_+_158519714
|
2.495
|
NM_001167903
NM_022736
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chrX_-_34675366
|
2.492
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr8_+_98788014
|
2.486
|
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr11_+_95523809
|
2.486
|
|
CEP57
|
centrosomal protein 57kDa
|
|
chr11_+_63706433
|
2.480
|
NM_024771
|
NAA40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae)
|
|
chr11_+_18344113
|
2.480
|
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr6_-_80656870
|
2.470
|
NM_022726
|
ELOVL4
|
ELOVL fatty acid elongase 4
|
|
chr10_+_112327411
|
2.455
|
NM_005445
|
SMC3
|
structural maintenance of chromosomes 3
|
|
chr10_+_46222647
|
2.455
|
NM_001169106
NM_001169107
NM_015262
|
FAM21C
|
family with sequence similarity 21, member C
|
|
chr6_-_31830551
|
2.447
|
|
NEU1
|
sialidase 1 (lysosomal sialidase)
|
|
chr10_-_93392744
|
2.444
|
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr7_+_116312412
|
2.440
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chrX_+_118108595
|
2.427
|
NM_001031855
NM_024778
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr3_-_126076057
|
2.427
|
NM_014079
|
KLF15
|
Kruppel-like factor 15
|
|
chr10_+_51827654
|
2.423
|
NM_001005751
|
FAM21C
FAM21A
|
family with sequence similarity 21, member C
family with sequence similarity 21, member A
|
|
chr2_+_109204745
|
2.422
|
NM_004987
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr8_-_95274520
|
2.421
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr16_-_1525040
|
2.420
|
NM_001114331
NM_001287
|
CLCN7
|
chloride channel 7
|
|
chr16_-_23521794
|
2.418
|
|
GGA2
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 2
|
|
chr6_+_127587984
|
2.417
|
|
RNF146
|
ring finger protein 146
|
|
chr3_+_158519877
|
2.414
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr3_+_158519925
|
2.392
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr21_-_46237974
|
2.392
|
|
SUMO3
|
SMT3 suppressor of mif two 3 homolog 3 (S. cerevisiae)
|
|
chr18_-_46987016
|
2.385
|
NM_017653
|
DYM
|
dymeclin
|
|
chr11_+_9685650
|
2.363
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr16_-_23521803
|
2.357
|
NM_015044
|
GGA2
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 2
|
|
chr8_-_81083660
|
2.342
|
NM_001025253
NM_005079
|
TPD52
|
tumor protein D52
|
|
chr8_-_126104004
|
2.318
|
|
KIAA0196
|
KIAA0196
|
|
chr8_+_71581599
|
2.317
|
NM_001011720
|
XKR9
|
XK, Kell blood group complex subunit-related family, member 9
|
|
chr6_-_44225024
|
2.313
|
NM_178148
|
SLC35B2
|
solute carrier family 35, member B2
|
|
chr7_+_155089614
|
2.313
|
|
INSIG1
|
insulin induced gene 1
|
|
chr7_-_17980102
|
2.299
|
NM_015132
|
SNX13
|
sorting nexin 13
|
|
chr9_-_125667493
|
2.295
|
NM_001100588
NM_018835
|
RC3H2
|
ring finger and CCCH-type domains 2
|
|
chr1_-_156265395
|
2.293
|
NM_144580
|
C1orf85
|
chromosome 1 open reading frame 85
|
|
chr17_-_7137675
|
2.292
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr19_-_19051112
|
2.291
|
NM_001145722
NM_001145721
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chr2_+_228189919
|
2.291
|
|
MFF
|
mitochondrial fission factor
|
|
chr8_-_126103926
|
2.290
|
|
KIAA0196
|
KIAA0196
|
|
chr5_+_43603312
|
2.282
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr2_-_20251788
|
2.280
|
NM_014713
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr12_+_72233486
|
2.279
|
NM_001146213
NM_001146214
NM_022771
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr1_+_76540374
|
2.272
|
NM_001160011
NM_152996
|
ST6GALNAC3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3
|
|
chr7_+_16793320
|
2.269
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chr15_-_72668301
|
2.265
|
NM_000520
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr19_+_5681016
|
2.254
|
NM_198704
NM_198705
NM_198533
NM_198707
NM_198706
NM_198708
|
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like
|
|
chr7_+_12726910
|
2.247
|
NM_001195396
NM_005738
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr16_-_1877140
|
2.243
|
NM_001040427
NM_005326
|
HAGH
|
hydroxyacylglutathione hydrolase
|
|
chr1_-_154531083
|
2.239
|
NM_017582
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr11_+_9685624
|
2.236
|
NM_015055
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|