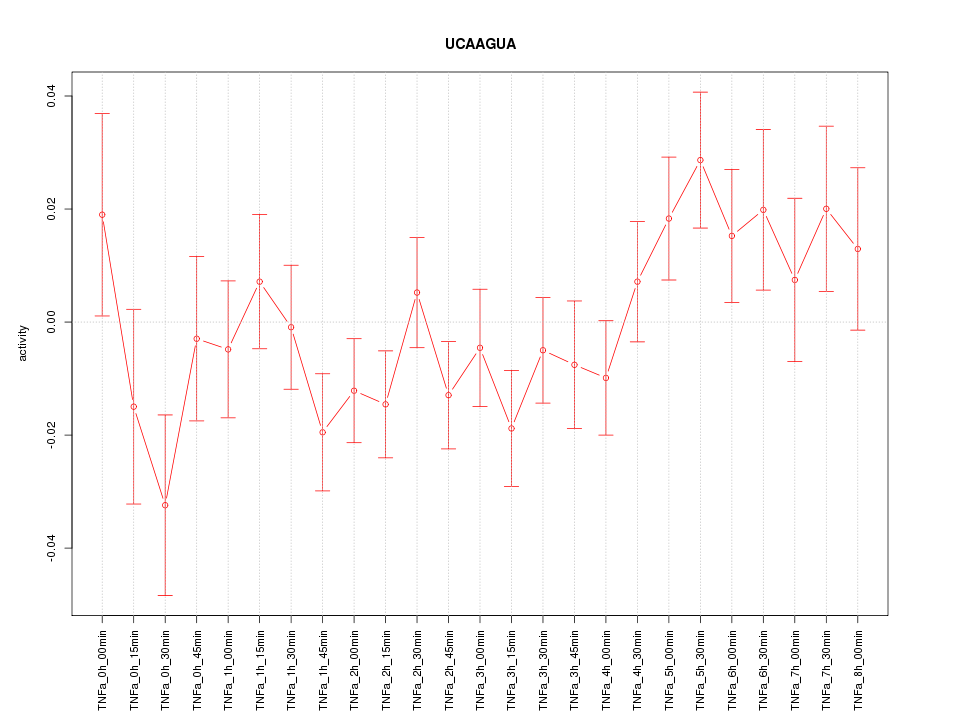
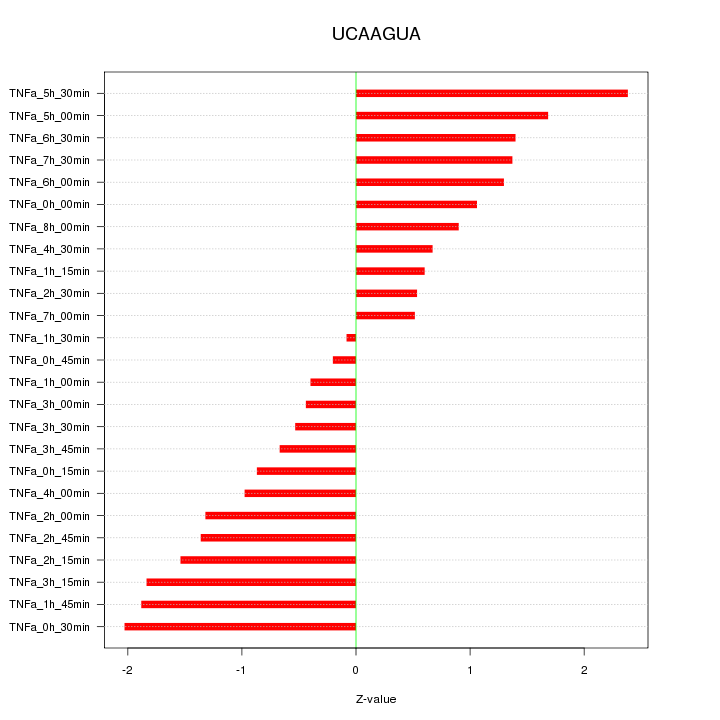
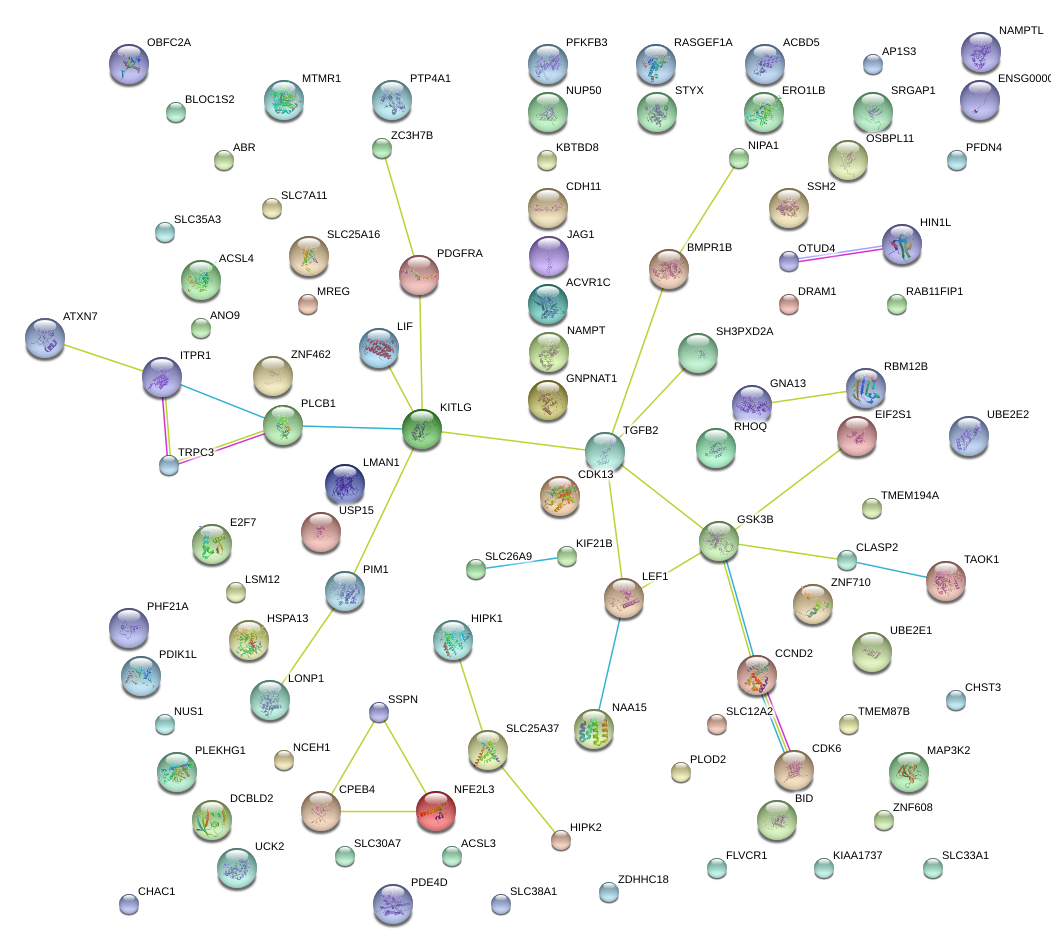
|
chr3_-_172428773
|
2.501
|
NM_001146276
NM_001146277
NM_001146278
NM_020792
|
NCEH1
|
neutral cholesterol ester hydrolase 1
|
|
chr2_+_223725675
|
2.258
|
NM_004457
NM_203372
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3
|
|
chr10_-_105614952
|
2.143
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr16_-_65155881
|
2.033
|
NM_001797
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr15_+_90544608
|
1.936
|
NM_198526
|
ZNF710
|
zinc finger protein 710
|
|
chr5_-_59189620
|
1.816
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_-_145879222
|
1.802
|
NM_000935
NM_182943
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr8_+_23386362
|
1.772
|
NM_016612
|
SLC25A37
|
solute carrier family 25, member 37
|
|
chr9_+_109625347
|
1.671
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr5_+_127419407
|
1.622
|
NM_001046
|
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 2
|
|
chr2_-_216878314
|
1.621
|
NM_018000
|
MREG
|
melanoregulin
|
|
chr5_-_59783885
|
1.576
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_-_57472573
|
1.568
|
NM_001130963
NM_015257
|
TMEM194A
|
transmembrane protein 194A
|
|
chr5_-_58571916
|
1.542
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_58295758
|
1.529
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_58334936
|
1.523
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_58882274
|
1.515
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_59064437
|
1.466
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_58652671
|
1.443
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_-_200992824
|
1.442
|
NM_001252100
NM_001252102
NM_001252103
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr12_+_102271082
|
1.396
|
NM_018370
|
DRAM1
|
DNA-damage regulated autophagy modulator 1
|
|
chr22_+_41697452
|
1.361
|
NM_017590
|
ZC3H7B
|
zinc finger CCCH-type containing 7B
|
|
chr7_-_92465929
|
1.353
|
NM_001145306
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr4_+_140222668
|
1.272
|
NM_057175
|
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit
|
|
chr3_-_98620452
|
1.257
|
NM_080927
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr3_+_23244703
|
1.255
|
NM_152653
|
UBE2E2
|
ubiquitin-conjugating enzyme E2E 2
|
|
chr7_-_92463211
|
1.252
|
NM_001259
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr1_+_101361589
|
1.207
|
NM_001144884
NM_133496
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr7_-_139477437
|
1.193
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr1_+_165796652
|
1.178
|
NM_012474
|
UCK2
|
uridine-cytidine kinase 2
|
|
chr8_-_37756971
|
1.157
|
NM_001002814
NM_025151
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chr18_-_57026465
|
1.140
|
NM_005570
|
LMAN1
|
lectin, mannose-binding, 1
|
|
chr15_-_23086290
|
1.140
|
NM_144599
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chrX_-_108976509
|
1.110
|
NM_004458
NM_022977
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr15_-_23086842
|
1.081
|
NM_001142275
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr2_+_112812799
|
1.080
|
NM_032824
|
TMEM87B
|
transmembrane protein 87B
|
|
chr12_+_64238511
|
1.077
|
NM_020762
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1
|
|
chr10_+_73723992
|
1.072
|
NM_004273
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3
|
|
chr4_-_139163222
|
1.042
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr15_+_41245635
|
1.018
|
NM_001142776
NM_024111
|
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli)
|
|
chr11_-_441995
|
0.994
|
NM_001012302
|
ANO9
|
anoctamin 9
|
|
chr20_+_8112762
|
0.945
|
NM_015192
NM_182734
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific)
|
|
chr22_+_45560529
|
0.944
|
NM_153645
|
NUP50
|
nucleoporin 50kDa
|
|
chr20_-_10654429
|
0.929
|
NM_000214
|
JAG1
|
jagged 1
|
|
chr6_+_117996616
|
0.919
|
NM_138459
|
NUS1
|
nuclear undecaprenyl pyrophosphate synthase 1 homolog (S. cerevisiae)
|
|
chr14_-_53258329
|
0.904
|
NM_198066
|
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1
|
|
chr4_-_122872908
|
0.863
|
NM_001130698
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3
|
|
chr17_+_27717942
|
0.854
|
NM_020791
NM_025142
|
TAOK1
|
TAO kinase 1
|
|
chr17_-_63052884
|
0.844
|
NM_006572
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr12_+_62654120
|
0.839
|
NM_001252078
NM_001252079
NM_006313
|
USP15
|
ubiquitin specific peptidase 15
|
|
chr2_+_46769821
|
0.835
|
NM_012249
|
RHOQ
|
ras homolog gene family, member Q
|
|
chr3_-_119812973
|
0.833
|
NM_001146156
NM_002093
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr1_+_218518675
|
0.823
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr3_-_125314367
|
0.811
|
NM_022776
|
OSBPL11
|
oxysterol binding protein-like 11
|
|
chr17_-_1083008
|
0.786
|
NM_021962
|
ABR
|
active BCR-related gene
|
|
chr22_+_45559725
|
0.776
|
NM_007172
|
NUP50
|
nucleoporin 50kDa
|
|
chr6_+_37137882
|
0.767
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr7_+_26191732
|
0.759
|
NM_004289
|
NFE2L3
|
nuclear factor (erythroid-derived 2)-like 3
|
|
chr2_+_192542860
|
0.733
|
NM_001031716
|
OBFC2A
|
oligonucleotide/oligosaccharide-binding fold containing 2A
|
|
chr12_+_4382882
|
0.721
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr12_+_26348268
|
0.714
|
NM_001135823
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr4_-_109089444
|
0.707
|
NM_001130713
NM_001130714
NM_016269
|
LEF1
|
lymphoid enhancer-binding factor 1
|
|
chr10_-_102046415
|
0.706
|
NM_173809
|
BLOC1S2
|
biogenesis of lysosomal organelles complex-1, subunit 2
|
|
chr10_-_102045948
|
0.705
|
NM_001001342
|
BLOC1S2
|
biogenesis of lysosomal organelles complex-1, subunit 2
|
|
chr5_+_173315313
|
0.705
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr3_+_67048726
|
0.686
|
NM_032505
|
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8
|
|
chr5_-_124080773
|
0.682
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr12_-_88974094
|
0.680
|
NM_000899
NM_003994
|
KITLG
|
KIT ligand
|
|
chr4_-_146100793
|
0.677
|
NM_001102653
|
OTUD4
|
OTU domain containing 4
|
|
chr4_-_109087876
|
0.667
|
NM_001166119
|
LEF1
|
lymphoid enhancer-binding factor 1
|
|
chr8_-_94753211
|
0.663
|
NM_203390
|
RBM12B
|
RNA binding motif protein 12B
|
|
chr4_-_122854192
|
0.662
|
NM_003305
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3
|
|
chr21_-_15755439
|
0.661
|
NM_006948
|
HSPA13
|
heat shock protein 70kDa family, member 13
|
|
chr14_+_67827030
|
0.657
|
NM_004094
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa
|
|
chr3_+_4535030
|
0.655
|
NM_001099952
NM_001168272
NM_002222
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1
|
|
chr10_+_6244839
|
0.654
|
NM_004566
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr3_-_155572166
|
0.651
|
NM_001190992
NM_004733
|
SLC33A1
|
solute carrier family 33 (acetyl-CoA transporter), member 1
|
|
chr4_+_55095263
|
0.638
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr10_-_70287249
|
0.636
|
NM_152707
|
SLC25A16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16
|
|
chr10_-_27529263
|
0.633
|
NM_145698
|
ACBD5
|
acyl-CoA binding domain containing 5
|
|
chr2_-_224702300
|
0.632
|
NM_001039569
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit
|
|
chr10_+_6186758
|
0.631
|
NM_001145443
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr6_+_150920998
|
0.629
|
NM_001029884
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1
|
|
chr22_-_18256748
|
0.626
|
NM_001244572
NM_197966
|
BID
|
BH3 interacting domain death agonist
|
|
chr1_+_100435539
|
0.620
|
NM_012243
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3
|
|
chr7_+_39989955
|
0.619
|
NM_003718
NM_031267
|
CDK13
|
cyclin-dependent kinase 13
|
|
chr12_-_46663207
|
0.614
|
NM_001077484
NM_030674
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr3_-_33700932
|
0.610
|
NM_001207044
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr17_-_1012257
|
0.601
|
NM_001092
|
ABR
|
active BCR-related gene
|
|
chr17_-_28257017
|
0.601
|
NM_033389
|
SSH2
|
slingshot homolog 2 (Drosophila)
|
|
chr14_+_53196852
|
0.601
|
NM_001130701
NM_145251
|
STYX
|
serine/threonine/tyrosine interacting protein
|
|
chr22_-_30642683
|
0.600
|
NM_002309
|
LIF
|
leukemia inhibitory factor (cholinergic differentiation factor)
|
|
chr12_-_77459198
|
0.597
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr2_-_128100673
|
0.581
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr1_+_27153130
|
0.571
|
NM_032283
|
ZDHHC18
|
zinc finger, DHHC-type containing 18
|
|
chr4_+_95679075
|
0.570
|
NM_001203
|
BMPR1B
|
bone morphogenetic protein receptor, type IB
|
|
chr7_-_105925410
|
0.569
|
NM_005746
|
NAMPT
|
nicotinamide phosphoribosyltransferase
|
|
chr14_+_77564570
|
0.569
|
NM_033426
|
KIAA1737
|
KIAA1737
|
|
chr17_-_1090562
|
0.560
|
NM_001159746
|
ABR
|
active BCR-related gene
|
|
chr11_-_46142955
|
0.559
|
NM_001101802
NM_016621
|
PHF21A
|
PHD finger protein 21A
|
|
chr17_-_42144986
|
0.556
|
NM_152344
|
LSM12
|
LSM12 homolog (S. cerevisiae)
|
|
chr1_-_205912546
|
0.552
|
NM_052934
NM_134325
|
SLC26A9
|
solute carrier family 26, member 9
|
|
chr1_+_26438267
|
0.551
|
NM_001243533
NM_152835
|
PDIK1L
|
PDLIM1 interacting kinase 1 like
|
|
chr1_+_114493766
|
0.551
|
NM_198269
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr2_-_158485393
|
0.547
|
NM_001111032
NM_001111033
NM_145259
|
ACVR1C
|
activin A receptor, type IC
|
|
chr10_-_43762366
|
0.546
|
NM_145313
|
RASGEF1A
|
RasGEF domain family, member 1A
|
|
chr1_+_213031596
|
0.543
|
NM_014053
|
FLVCR1
|
feline leukemia virus subgroup C cellular receptor 1
|
|
chr6_+_64281910
|
0.540
|
NM_003463
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chrX_+_149861830
|
0.540
|
NM_003828
|
MTMR1
|
myotubularin related protein 1
|
|
chr20_+_52824393
|
0.536
|
NM_002623
|
PFDN4
|
prefoldin subunit 4
|
|
chr1_-_236445294
|
0.530
|
NM_019891
|
ERO1LB
|
ERO1-like beta (S. cerevisiae)
|
|
chr3_+_63850181
|
0.526
|
NM_000333
|
ATXN7
|
ataxin 7
|
|
chr10_-_118765087
|
0.524
|
NM_001127211
NM_018330
|
KIAA1598
|
KIAA1598
|
|
chr12_+_12764755
|
0.523
|
NM_001310
|
CREBL2
|
cAMP responsive element binding protein-like 2
|
|
chr12_-_133464197
|
0.523
|
NM_001161344
NM_001161345
NM_001161346
NM_001161347
NM_018223
|
CHFR
|
checkpoint with forkhead and ring finger domains
|
|
chr11_-_89956531
|
0.519
|
NM_001144073
NM_012124
|
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1
|
|
chr2_-_158454025
|
0.514
|
NM_001111031
|
ACVR1C
|
activin A receptor, type IC
|
|
chr11_-_57092283
|
0.513
|
NM_033396
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa
|
|
chr7_+_7222142
|
0.511
|
NM_020156
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1
|
|
chr2_-_33824294
|
0.510
|
NM_015475
|
FAM98A
|
family with sequence similarity 98, member A
|
|
chr15_+_89631380
|
0.508
|
NM_007011
NM_152924
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr12_+_26348488
|
0.506
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr12_+_65218351
|
0.503
|
NM_015279
|
TBC1D30
|
TBC1 domain family, member 30
|
|
chr5_+_89770680
|
0.501
|
NM_006467
|
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)
|
|
chr18_-_25757351
|
0.500
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr11_-_16424391
|
0.500
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr7_-_151329153
|
0.499
|
NM_024429
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit
|
|
chr12_-_26985922
|
0.498
|
NM_002223
|
ITPR2
|
inositol 1,4,5-trisphosphate receptor, type 2
|
|
chr16_+_88493878
|
0.497
|
NM_001127464
|
ZNF469
|
zinc finger protein 469
|
|
chr4_+_57774035
|
0.496
|
NM_005612
|
REST
|
RE1-silencing transcription factor
|
|
chr3_-_150264280
|
0.492
|
NM_014445
|
SERP1
|
stress-associated endoplasmic reticulum protein 1
|
|
chr11_+_36317724
|
0.490
|
NM_001160167
|
PRR5L
|
proline rich 5 like
|
|
chr10_+_127408083
|
0.489
|
NM_001202438
NM_015608
|
C10orf137
|
chromosome 10 open reading frame 137
|
|
chr3_+_63884074
|
0.481
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr6_-_33548017
|
0.480
|
NM_001188
|
BAK1
|
BCL2-antagonist/killer 1
|
|
chr10_+_18549583
|
0.479
|
NM_000724
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr12_-_63328663
|
0.477
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr1_+_114496498
|
0.476
|
NM_181358
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr8_+_86157693
|
0.476
|
NM_198584
|
CA13
|
carbonic anhydrase XIII
|
|
chr1_-_16482426
|
0.475
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr22_-_18257238
|
0.474
|
NM_001196
NM_001244569
NM_001244570
NM_197967
NM_001244567
|
BID
|
BH3 interacting domain death agonist
|
|
chr6_+_131456825
|
0.474
|
NM_016377
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chr3_+_142838596
|
0.472
|
NM_004267
|
CHST2
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2
|
|
chr16_-_89556886
|
0.471
|
NM_013275
|
ANKRD11
|
ankyrin repeat domain 11
|
|
chr20_+_43595119
|
0.470
|
NM_006282
|
STK4
|
serine/threonine kinase 4
|
|
chr2_-_69614321
|
0.465
|
NM_001244710
NM_002056
|
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1
|
|
chr1_-_23857711
|
0.464
|
NM_004091
|
E2F2
|
E2F transcription factor 2
|
|
chr6_+_70576447
|
0.463
|
NM_001858
|
COL19A1
|
collagen, type XIX, alpha 1
|
|
chr4_-_76598289
|
0.459
|
NM_012297
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr13_+_26828755
|
0.456
|
NM_001260
|
CDK8
|
cyclin-dependent kinase 8
|
|
chr10_+_60094712
|
0.454
|
NM_001204880
NM_003338
|
UBE2D1
|
ubiquitin-conjugating enzyme E2D 1
|
|
chr4_+_57775064
|
0.451
|
NM_001193508
|
REST
|
RE1-silencing transcription factor
|
|
chr5_+_60628085
|
0.450
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr8_-_23261675
|
0.449
|
NM_002318
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr4_-_76598611
|
0.448
|
NM_203504
NM_203505
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr14_+_92789536
|
0.448
|
NM_153647
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr4_+_72052354
|
0.448
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr1_+_160370363
|
0.447
|
NM_020335
|
VANGL2
|
vang-like 2 (van gogh, Drosophila)
|
|
chr5_+_71403040
|
0.434
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr3_+_173116243
|
0.432
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr10_+_65281108
|
0.431
|
NM_001001330
|
REEP3
|
receptor accessory protein 3
|
|
chr7_-_27205148
|
0.431
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr8_-_122653493
|
0.429
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr7_-_156803224
|
0.426
|
NM_005515
|
MNX1
|
motor neuron and pancreas homeobox 1
|
|
chr15_+_52311408
|
0.425
|
NM_002748
|
MAPK6
|
mitogen-activated protein kinase 6
|
|
chr10_+_86088156
|
0.423
|
NM_018999
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr2_-_7005764
|
0.419
|
NM_207315
|
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial
|
|
chr10_-_60027630
|
0.418
|
NM_152230
|
IPMK
|
inositol polyphosphate multikinase
|
|
chr19_+_19322763
|
0.415
|
NM_004386
|
NCAN
|
neurocan
|
|
chr4_-_170924911
|
0.415
|
NM_001009554
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr1_-_202679415
|
0.414
|
NM_177402
|
SYT2
|
synaptotagmin II
|
|
chr11_-_10830581
|
0.413
|
NM_001042559
NM_001418
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2
|
|
chr3_-_167452593
|
0.412
|
NM_007217
NM_145860
NM_145859
|
PDCD10
|
programmed cell death 10
|
|
chr5_+_34929679
|
0.411
|
NM_001012339
NM_194283
|
DNAJC21
|
DnaJ (Hsp40) homolog, subfamily C, member 21
|
|
chr1_-_212588239
|
0.410
|
NM_001198862
NM_018252
|
TMEM206
|
transmembrane protein 206
|
|
chr2_+_162016926
|
0.409
|
NM_001199135
|
TANK
|
TRAF family member-associated NFKB activator
|
|
chr10_-_27531041
|
0.408
|
NM_001042473
|
ACBD5
|
acyl-CoA binding domain containing 5
|
|
chr8_-_74791098
|
0.405
|
NM_001001481
NM_018299
|
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative)
|
|
chr1_-_169455099
|
0.402
|
NM_006996
|
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2
|
|
chr6_-_136788012
|
0.401
|
NM_001198615
|
MAP7
|
microtubule-associated protein 7
|
|
chr16_-_47007567
|
0.398
|
NM_005880
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2
|
|
chr12_-_44200047
|
0.395
|
NM_001242397
NM_002822
|
TWF1
|
twinfilin, actin-binding protein, homolog 1 (Drosophila)
|
|
chr3_+_63953419
|
0.392
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr12_+_65004191
|
0.389
|
NM_178169
|
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3
|
|
chr14_-_45431121
|
0.389
|
NM_017658
|
KLHL28
|
kelch-like 28 (Drosophila)
|
|
chr5_-_131132715
|
0.386
|
NM_001008738
NM_133372
|
FNIP1
|
folliculin interacting protein 1
|
|
chr7_+_91875411
|
0.384
|
NM_019004
|
ANKIB1
|
ankyrin repeat and IBR domain containing 1
|
|
chr13_-_30169795
|
0.383
|
NM_003045
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1
|
|
chr9_-_35115844
|
0.381
|
NM_025182
|
FAM214B
|
family with sequence similarity 214, member B
|
|
chr2_+_54785453
|
0.380
|
NM_178313
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr1_+_244214552
|
0.380
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr6_-_136871956
|
0.372
|
NM_001198616
NM_001198617
NM_003980
|
MAP7
|
microtubule-associated protein 7
|
|
chr16_+_30935895
|
0.372
|
NM_001099784
|
FBXL19
|
F-box and leucine-rich repeat protein 19
|
|
chr21_-_16437125
|
0.371
|
NM_003489
|
NRIP1
|
nuclear receptor interacting protein 1
|
|
chr5_-_56247915
|
0.371
|
NM_152622
|
MIER3
|
mesoderm induction early response 1, family member 3
|
|
chr6_-_88875766
|
0.371
|
NM_001160226
NM_001160258
NM_001160259
NM_016083
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr11_+_109964086
|
0.369
|
NM_033390
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr22_-_42017032
|
0.367
|
NM_015704
|
PPPDE2
|
PPPDE peptidase domain containing 2
|
|
chr1_+_70876863
|
0.366
|
NM_001190463
NM_001902
NM_153742
|
CTH
|
cystathionase (cystathionine gamma-lyase)
|
|
chr18_+_29672568
|
0.365
|
NM_001191324
NM_198128
|
RNF138
|
ring finger protein 138
|