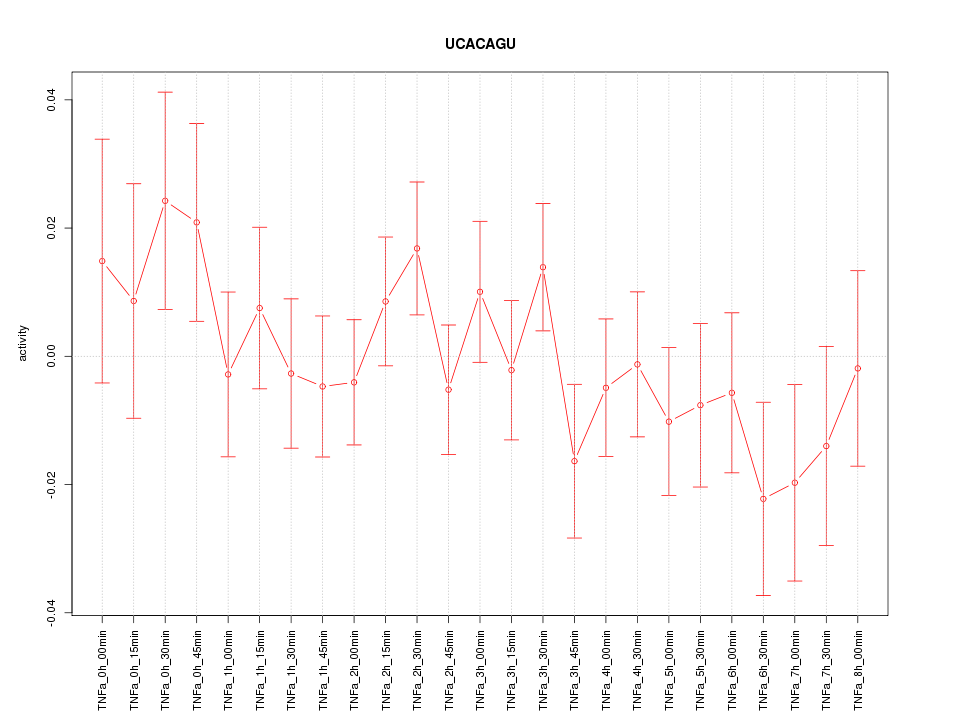
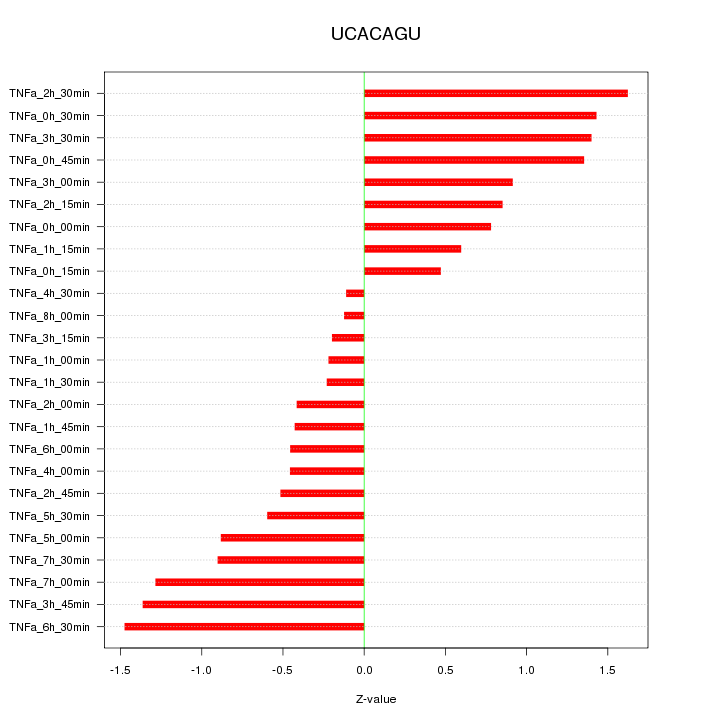
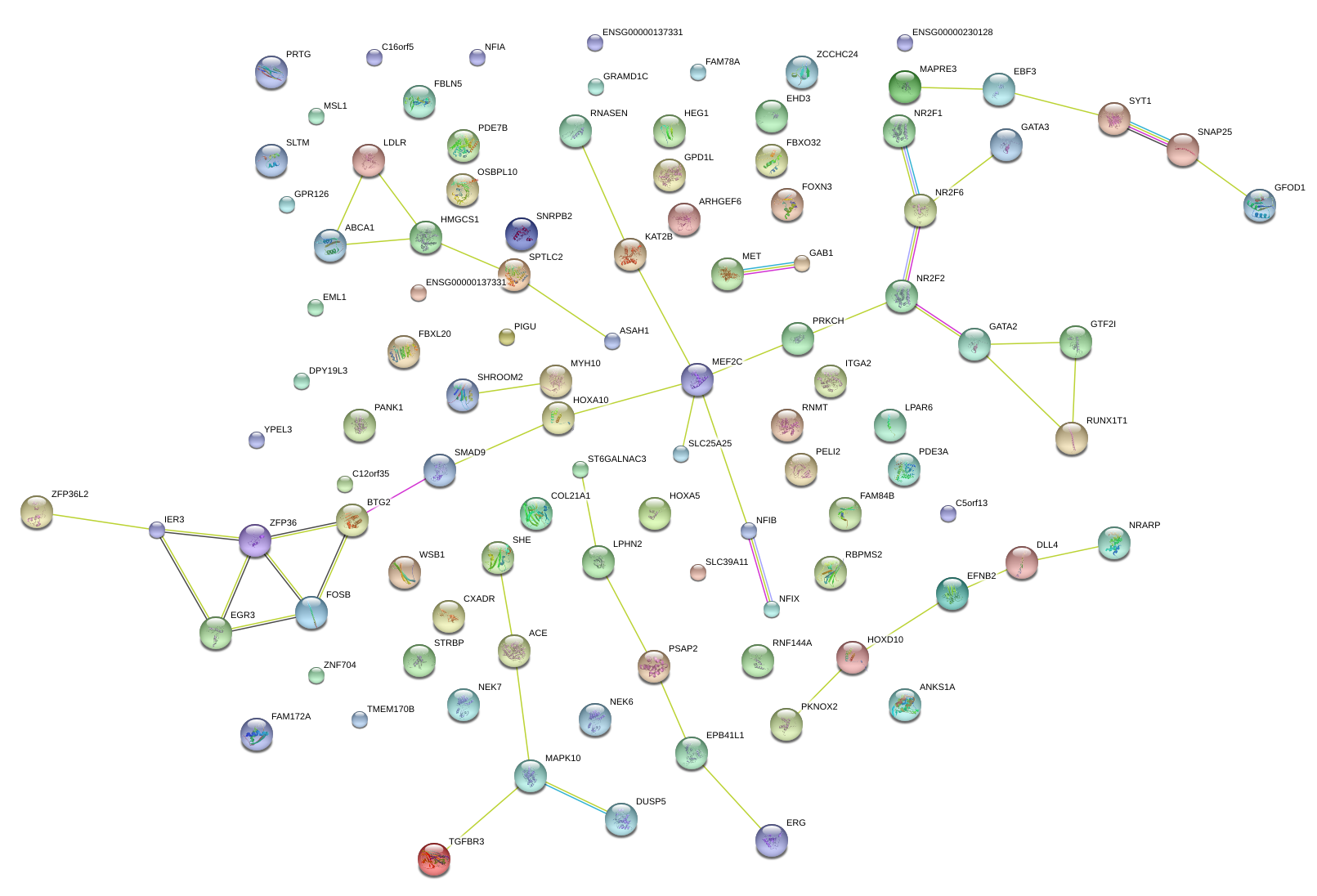
|
chr1_-_92371558
|
3.507
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr1_-_92351613
|
3.368
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr1_+_61542928
|
2.952
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr2_+_7057471
|
2.890
|
NM_014746
|
RNF144A
|
ring finger protein 144A
|
|
chr19_+_39897457
|
2.838
|
NM_003407
|
ZFP36
|
zinc finger protein 36, C3H type, homolog (mouse)
|
|
chr1_+_61547625
|
2.824
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr6_-_13408368
|
2.814
|
NM_001242630
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr10_-_131762017
|
2.728
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr6_+_142622797
|
2.668
|
NM_001032394
NM_001032395
NM_020455
NM_198569
|
GPR126
|
G protein-coupled receptor 126
|
|
chr1_+_61547388
|
2.605
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr10_-_81205091
|
2.400
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr9_-_107690526
|
2.313
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr9_-_134151905
|
2.233
|
NM_033387
|
FAM78A
|
family with sequence similarity 78, member A
|
|
chr9_-_14398981
|
2.175
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr1_-_154474524
|
2.163
|
NM_001010846
|
SHE
|
Src homology 2 domain containing E
|
|
chr8_-_127570710
|
2.054
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr1_+_203274646
|
2.003
|
NM_006763
|
BTG2
|
BTG family, member 2
|
|
chr9_-_14314036
|
1.982
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr14_+_56584854
|
1.891
|
NM_021255
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr7_-_27183225
|
1.866
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr21_+_18885104
|
1.817
|
NM_001207063
NM_001207064
NM_001207065
NM_001207066
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr7_-_27213873
|
1.810
|
NM_018951
|
HOXA10
|
homeobox A10
|
|
chr8_-_93029864
|
1.806
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_22549901
|
1.803
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr4_+_144257982
|
1.802
|
NM_002039
NM_207123
|
GAB1
|
GRB2-associated binding protein 1
|
|
chr8_-_22550708
|
1.781
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr17_+_25621105
|
1.781
|
NM_015626
NM_134265
|
WSB1
|
WD repeat and SOCS box containing 1
|
|
chr19_+_45971249
|
1.762
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr8_-_22550057
|
1.741
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr12_+_20522178
|
1.740
|
NM_000921
|
PDE3A
|
phosphodiesterase 3A, cGMP-inhibited
|
|
chr8_-_93107881
|
1.738
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr9_-_140196702
|
1.736
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr6_+_11538486
|
1.732
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr8_-_93111915
|
1.706
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr5_-_111093251
|
1.695
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr22_+_51113069
|
1.664
|
NM_001080420
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3
|
|
chr8_-_93107442
|
1.642
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr13_-_49001042
|
1.635
|
NM_001162497
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr11_+_125034558
|
1.619
|
NM_022062
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr5_-_111093030
|
1.617
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr7_-_28998028
|
1.596
|
NM_014817
|
TRIL
|
TLR4 interactor with leucine-rich repeats
|
|
chr3_+_32148000
|
1.579
|
NM_015141
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr1_+_82266081
|
1.547
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr5_-_111093792
|
1.514
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr8_-_93115453
|
1.495
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr17_+_61562177
|
1.488
|
NM_001178057
NM_152830
|
ACE
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1
|
|
chr5_-_111092918
|
1.484
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr2_+_31456879
|
1.468
|
NM_014600
|
EHD3
|
EH-domain containing 3
|
|
chr17_+_61554420
|
1.448
|
NM_000789
|
ACE
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1
|
|
chr5_-_111312522
|
1.417
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr6_+_136172802
|
1.399
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr9_+_130830447
|
1.383
|
NM_001006641
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr9_+_130854127
|
1.375
|
NM_001006642
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr9_+_130860816
|
1.372
|
NM_052901
NM_001006643
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr14_-_92414004
|
1.345
|
NM_006329
|
FBLN5
|
fibulin 5
|
|
chr19_+_32897021
|
1.341
|
NM_207325
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr13_-_48987652
|
1.338
|
NM_001162498
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr3_+_20081523
|
1.305
|
NM_003884
|
KAT2B
|
K(lysine) acetyltransferase 2B
|
|
chr19_+_32896515
|
1.298
|
NM_001172774
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr8_-_93075190
|
1.295
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_124544376
|
1.291
|
NM_148177
|
FBXO32
|
F-box protein 32
|
|
chr16_-_4588379
|
1.273
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr20_+_34742656
|
1.273
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr14_+_61788398
|
1.242
|
NM_006255
|
PRKCH
|
protein kinase C, eta
|
|
chr5_-_111091947
|
1.232
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr6_-_30712294
|
1.225
|
NM_003897
|
IER3
|
immediate early response 3
|
|
chr8_-_124553448
|
1.223
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr14_-_78083109
|
1.221
|
NM_004863
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2
|
|
chr17_-_71088832
|
1.202
|
NM_001159770
NM_139177
|
SLC39A11
|
solute carrier family 39 (metal ion transporter), member 11
|
|
chr3_-_128212027
|
1.192
|
NM_032638
|
GATA2
|
GATA binding protein 2
|
|
chr17_-_8534008
|
1.191
|
NM_005964
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr9_+_127054246
|
1.175
|
NM_001166170
NM_001166171
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr3_-_124774735
|
1.169
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr15_+_96876568
|
1.151
|
NM_001145157
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr15_-_65067696
|
1.141
|
NM_194272
|
RBPMS2
|
RNA binding protein with multiple splicing 2
|
|
chr13_-_37494370
|
1.136
|
NM_001127217
NM_005905
|
SMAD9
|
SMAD family member 9
|
|
chr5_-_31532164
|
1.100
|
NM_001100412
NM_013235
|
DROSHA
|
drosha, ribonuclease type III
|
|
chr9_+_127054567
|
1.100
|
NM_001166169
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr13_-_107187312
|
1.096
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr12_+_79257772
|
1.095
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr14_+_100259445
|
1.090
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr20_+_34700257
|
1.090
|
NM_177996
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr6_-_13486382
|
1.086
|
NM_001242628
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr15_+_41221530
|
1.077
|
NM_019074
|
DLL4
|
delta-like 4 (Drosophila)
|
|
chr12_+_79439432
|
1.076
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr10_+_112257624
|
1.071
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr3_-_128207366
|
1.069
|
NM_001145661
|
GATA2
|
GATA binding protein 2
|
|
chr1_+_76540374
|
1.066
|
NM_001160011
NM_152996
|
ST6GALNAC3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3
|
|
chr16_-_4588738
|
1.056
|
NM_001199055
NM_001199056
NM_013399
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr5_-_88179278
|
1.042
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr15_+_96875793
|
1.028
|
NM_001145156
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr20_+_10199455
|
1.020
|
NM_003081
NM_130811
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr2_-_43453737
|
1.009
|
NM_006887
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr12_+_79258448
|
1.005
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr5_-_93447237
|
0.993
|
NM_001163417
NM_001163418
NM_032042
|
FAM172A
|
family with sequence similarity 172, member A
|
|
chr10_+_8096633
|
0.967
|
NM_001002295
NM_002051
|
GATA3
|
GATA binding protein 3
|
|
chr6_+_34857021
|
0.964
|
NM_015245
|
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A
|
|
chr7_+_116312412
|
0.962
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr15_+_96869156
|
0.948
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr9_+_127019782
|
0.941
|
NM_001166167
NM_001145001
NM_014397
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr5_-_88199868
|
0.938
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr16_-_30107492
|
0.912
|
NM_031477
NM_001145524
|
YPEL3
|
yippee-like 3 (Drosophila)
|
|
chrX_-_135863502
|
0.908
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr9_-_126030854
|
0.895
|
NM_001171137
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr19_-_17356022
|
0.894
|
NM_005234
|
NR2F6
|
nuclear receptor subfamily 2, group F, member 6
|
|
chr9_+_127020462
|
0.892
|
NM_001166168
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr15_-_56035176
|
0.870
|
NM_173814
|
PRTG
|
protogenin
|
|
chr7_+_74072007
|
0.864
|
NM_001163636
NM_001518
NM_032999
NM_033000
NM_033001
|
GTF2I
|
general transcription factor IIi
|
|
chr9_-_126030792
|
0.849
|
NM_018387
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr6_-_56112315
|
0.842
|
NM_030820
|
COL21A1
|
collagen, type XXI, alpha 1
|
|
chr8_-_81787015
|
0.803
|
NM_001033723
|
ZNF704
|
zinc finger protein 704
|
|
chr19_+_11200037
|
0.790
|
NM_000527
NM_001195798
NM_001195799
NM_001195800
NM_001195802
NM_001195803
|
LDLR
|
low density lipoprotein receptor
|
|
chr12_+_32112286
|
0.778
|
NM_018169
|
C12orf35
|
chromosome 12 open reading frame 35
|
|
chr5_+_52284903
|
0.775
|
NM_002203
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor)
|
|
chrX_+_9754465
|
0.766
|
NM_001649
|
SHROOM2
|
shroom family member 2
|
|
chr14_-_89883306
|
0.747
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr17_+_38278789
|
0.734
|
NM_001012241
|
MSL1
|
male-specific lethal 1 homolog (Drosophila)
|
|
chr15_+_96873845
|
0.727
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr5_-_88119604
|
0.705
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr8_-_17941617
|
0.688
|
NM_177924
|
ASAH1
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1
|
|
chr21_-_40033617
|
0.685
|
NM_001243432
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr3_-_32023237
|
0.682
|
NM_001174060
NM_017784
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr5_-_43313581
|
0.681
|
NM_001098272
NM_002130
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble)
|
|
chr10_-_91403630
|
0.679
|
NM_138316
NM_148978
|
PANK1
|
pantothenate kinase 1
|
|
chr17_-_37557851
|
0.673
|
NM_001184906
NM_032875
|
FBXL20
|
F-box and leucine-rich repeat protein 20
|
|
chr21_-_39870303
|
0.664
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr3_+_113557679
|
0.664
|
NM_017577
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr3_-_64211111
|
0.660
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chrX_-_67653169
|
0.653
|
NM_002547
|
OPHN1
|
oligophrenin 1
|
|
chr3_+_113616317
|
0.650
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr1_+_116185249
|
0.648
|
NM_001172412
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr5_-_73937248
|
0.639
|
NM_003633
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr8_-_17942410
|
0.636
|
NM_001127505
NM_004315
|
ASAH1
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1
|
|
chr19_-_36391448
|
0.631
|
NM_139239
|
NFKBID
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta
|
|
chr12_+_32654976
|
0.624
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr6_-_13487772
|
0.622
|
NM_001242629
NM_018988
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr21_-_44846927
|
0.619
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr5_+_176560056
|
0.614
|
NM_172349
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr1_-_36023005
|
0.610
|
NM_024874
|
KIAA0319L
|
KIAA0319-like
|
|
chr3_+_23987514
|
0.600
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr12_-_12419702
|
0.598
|
NM_002336
|
LRP6
|
low density lipoprotein receptor-related protein 6
|
|
chr10_+_70320044
|
0.597
|
NM_030625
|
TET1
|
tet methylcytosine dioxygenase 1
|
|
chr3_+_58223228
|
0.587
|
NM_020676
|
ABHD6
|
abhydrolase domain containing 6
|
|
chr11_-_67888857
|
0.580
|
NM_001277
NM_212469
|
CHKA
|
choline kinase alpha
|
|
chr11_+_118230292
|
0.571
|
NM_001204077
NM_004788
|
UBE4A
|
ubiquitination factor E4A
|
|
chrX_+_105066531
|
0.569
|
NM_198465
|
NRK
|
Nik related kinase
|
|
chr9_-_98269480
|
0.567
|
NM_001083607
|
PTCH1
|
patched 1
|
|
chr15_+_91447419
|
0.566
|
NM_006122
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2
|
|
chr18_-_72921280
|
0.564
|
NM_175907
|
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2
|
|
chr1_-_68299047
|
0.564
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr3_-_105587710
|
0.560
|
NM_170662
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr1_+_116184573
|
0.558
|
NM_001172411
NM_138959
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr9_-_98270321
|
0.558
|
NM_001083604
NM_001083605
|
PTCH1
|
patched 1
|
|
chrX_+_102632108
|
0.554
|
NM_014380
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr5_+_74632992
|
0.551
|
NM_000859
NM_001130996
|
HMGCR
|
3-hydroxy-3-methylglutaryl-CoA reductase
|
|
chr17_+_38474472
|
0.547
|
NM_001145301
NM_001145302
|
RARA
|
retinoic acid receptor, alpha
|
|
chr12_-_118541655
|
0.544
|
NM_019086
|
VSIG10
|
V-set and immunoglobulin domain containing 10
|
|
chr5_+_53813494
|
0.541
|
NM_001145427
NM_001102575
NM_052870
|
SNX18
|
sorting nexin 18
|
|
chr4_+_52709165
|
0.536
|
NM_001040402
NM_015115
|
DCUN1D4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae)
|
|
chr10_-_52383611
|
0.529
|
NM_147156
|
SGMS1
|
sphingomyelin synthase 1
|
|
chr20_-_33999780
|
0.528
|
NM_001184977
NM_018244
NM_199487
|
UQCC
|
ubiquinol-cytochrome c reductase complex chaperone
|
|
chr1_-_174992552
|
0.525
|
NM_022100
|
MRPS14
|
mitochondrial ribosomal protein S14
|
|
chr14_-_90085325
|
0.524
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr21_-_34100316
|
0.519
|
NM_003895
NM_203446
|
SYNJ1
|
synaptojanin 1
|
|
chr1_+_211433285
|
0.516
|
NM_018254
|
RCOR3
|
REST corepressor 3
|
|
chr12_+_83080901
|
0.516
|
NM_152588
|
TMTC2
|
transmembrane and tetratricopeptide repeat containing 2
|
|
chr9_-_98270830
|
0.515
|
NM_000264
|
PTCH1
|
patched 1
|
|
chr2_-_70475563
|
0.513
|
NM_022037
NM_022173
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein
|
|
chr15_-_37393364
|
0.510
|
NM_001220482
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr17_-_35969401
|
0.500
|
NM_001163544
NM_001163545
NM_001163546
NM_001163547
NM_007247
NM_080550
NM_198882
|
SYNRG
|
synergin, gamma
|
|
chr17_-_49337370
|
0.499
|
NM_017643
|
MBTD1
|
mbt domain containing 1
|
|
chrX_+_102631267
|
0.499
|
NM_206915
NM_206917
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr5_-_38595506
|
0.498
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr3_-_134093235
|
0.498
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr10_-_91405320
|
0.495
|
NM_148977
|
PANK1
|
pantothenate kinase 1
|
|
chr12_-_95044205
|
0.492
|
NM_020698
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chr4_-_114682836
|
0.491
|
NM_001221
NM_172114
NM_172115
NM_172127
NM_172128
NM_172129
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta
|
|
chr11_-_74109417
|
0.488
|
NM_173582
|
PGM2L1
|
phosphoglucomutase 2-like 1
|
|
chr2_+_61292999
|
0.484
|
NM_001129993
|
KIAA1841
|
KIAA1841
|
|
chr5_-_38556682
|
0.479
|
NM_001127671
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr12_-_65153189
|
0.479
|
NM_002076
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr2_+_113403433
|
0.476
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr4_+_26862312
|
0.475
|
NM_001169117
NM_001169118
NM_020860
|
STIM2
|
stromal interaction molecule 2
|
|
chr2_+_242641310
|
0.456
|
NM_032329
|
ING5
|
inhibitor of growth family, member 5
|
|
chr12_-_31743822
|
0.450
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr14_-_39901619
|
0.447
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr3_-_39195101
|
0.445
|
NM_033027
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1
|
|
chr21_+_35445808
|
0.445
|
NM_032476
NM_006933
|
MRPS6
SLC5A3
|
mitochondrial ribosomal protein S6
solute carrier family 5 (sodium/myo-inositol cotransporter), member 3
|
|
chr17_+_28705922
|
0.442
|
NM_001304
|
CPD
|
carboxypeptidase D
|
|
chr10_+_97803064
|
0.442
|
NM_001134375
NM_001134376
NM_019084
|
CCNJ
|
cyclin J
|
|
chr2_-_200335988
|
0.442
|
NM_015265
|
SATB2
|
SATB homeobox 2
|
|
chr7_+_94285676
|
0.440
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr10_+_35535952
|
0.440
|
NM_181698
|
CCNY
|
cyclin Y
|
|
chr15_-_37390372
|
0.436
|
NM_172315
|
MEIS2
|
Meis homeobox 2
|
|
chr8_-_57123858
|
0.430
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr3_+_98451537
|
0.426
|
NM_006100
|
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6
|
|
chr3_-_129599220
|
0.424
|
NM_001017395
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr1_-_225840660
|
0.422
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr17_+_28707495
|
0.418
|
NM_001199775
|
CPD
|
carboxypeptidase D
|
|
chr7_+_2552162
|
0.417
|
NM_001166355
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|