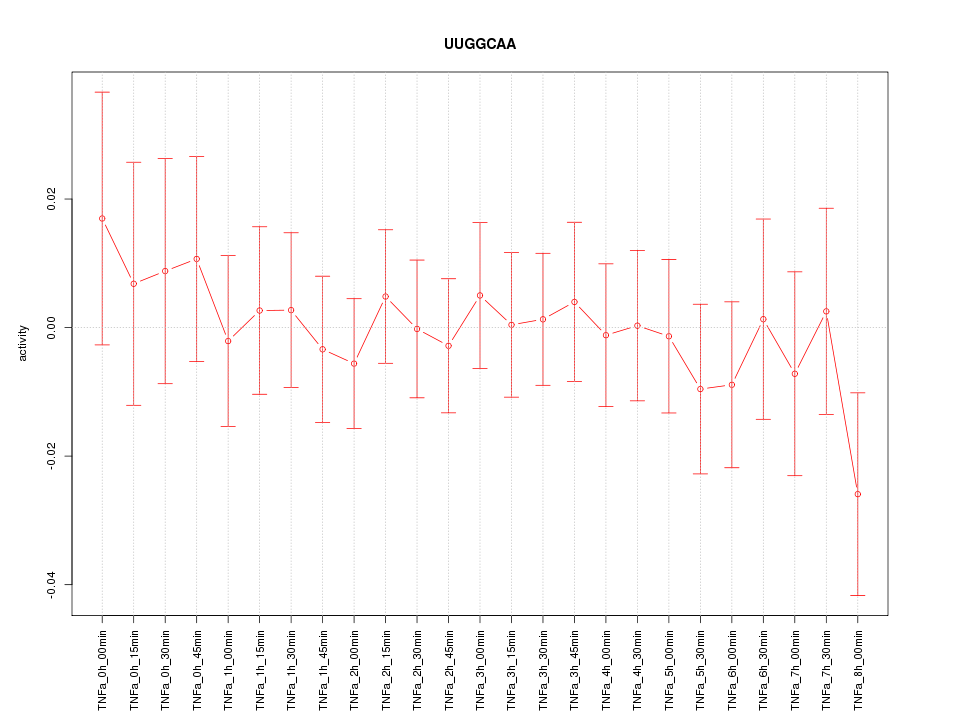
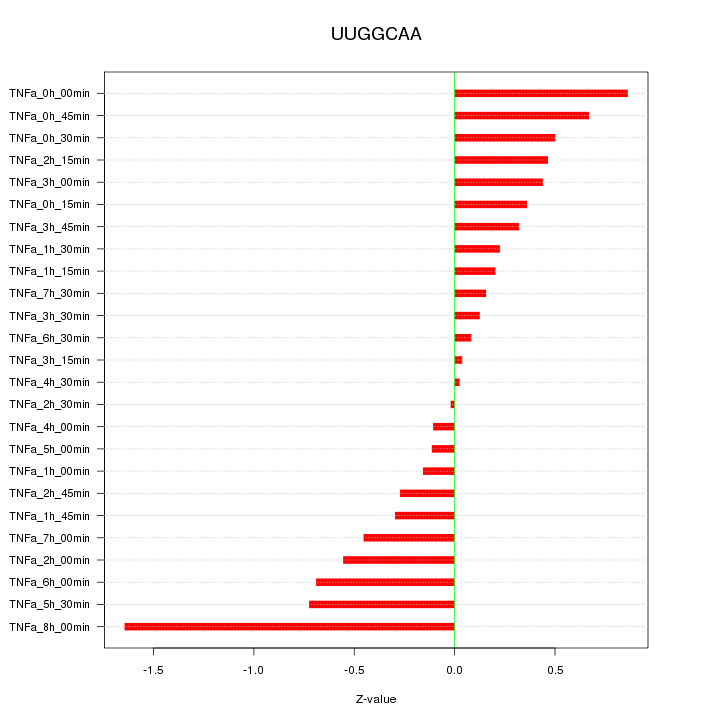
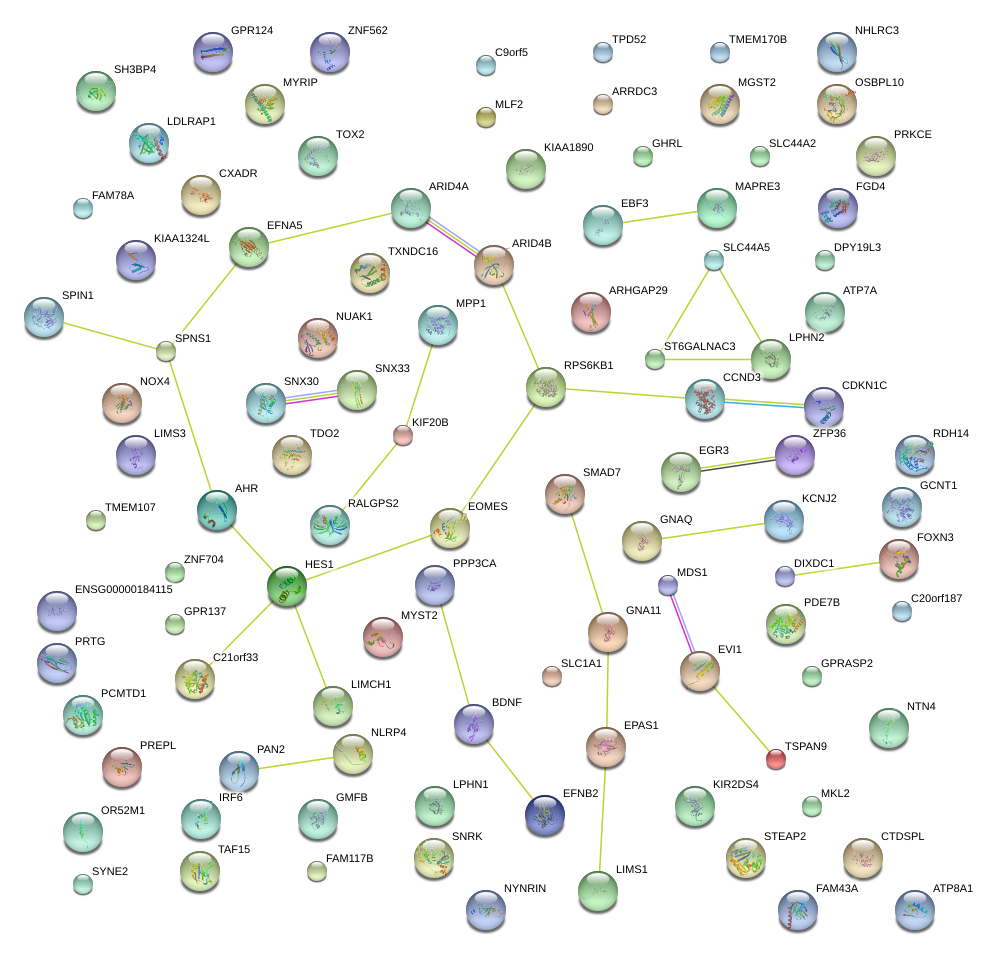
|
chr10_-_131762017
|
1.964
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr9_+_79056581
|
1.671
|
NM_001097634
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr9_+_79074067
|
1.581
|
NM_001097633
NM_001490
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr9_+_79093256
|
1.565
|
NM_001097635
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr3_+_193853930
|
1.554
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr9_+_79115548
|
1.546
|
NM_001097636
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr8_-_22550708
|
1.530
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr8_-_22549901
|
1.511
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr3_+_39851028
|
1.465
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr8_-_22550057
|
1.462
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr9_-_134151905
|
1.312
|
NM_033387
|
FAM78A
|
family with sequence similarity 78, member A
|
|
chr20_+_42544781
|
1.170
|
NM_001098796
NM_032883
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr8_-_80993009
|
1.165
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr8_-_81083660
|
1.156
|
NM_001025253
NM_005079
|
TPD52
|
tumor protein D52
|
|
chr20_+_42543417
|
1.107
|
NM_001098797
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr1_+_82266081
|
1.099
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr20_+_42574535
|
1.093
|
NM_001098798
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr4_-_102268626
|
1.072
|
NM_000944
NM_001130691
NM_001130692
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr17_+_68165660
|
1.060
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr21_+_18885104
|
1.055
|
NM_001207063
NM_001207064
NM_001207065
NM_001207066
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chrX_+_101967103
|
0.989
|
NM_001004051
NM_001184874
NM_001184875
NM_001184876
NM_138437
|
GPRASP2
|
G protein-coupled receptor associated sorting protein 2
|
|
chr12_-_106533810
|
0.981
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr12_+_32654976
|
0.956
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr4_+_41362750
|
0.945
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr1_-_209979388
|
0.926
|
NM_001206696
NM_006147
|
IRF6
|
interferon regulatory factor 6
|
|
chr4_+_41614911
|
0.923
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr18_-_46475702
|
0.911
|
NM_001190822
|
SMAD7
|
SMAD family member 7
|
|
chr8_+_37654395
|
0.896
|
NM_032777
|
GPR124
|
G protein-coupled receptor 124
|
|
chr7_+_17338182
|
0.873
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr3_-_168865521
|
0.869
|
NM_001164000
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr3_+_37903643
|
0.860
|
NM_001008392
NM_005808
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chr14_+_64682671
|
0.856
|
NM_182910
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr18_-_46477080
|
0.851
|
NM_001190821
NM_005904
|
SMAD7
|
SMAD family member 7
|
|
chr3_-_168864325
|
0.847
|
NM_001105078
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr14_+_64680858
|
0.846
|
NM_182913
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr3_-_168864066
|
0.843
|
NM_001105077
NM_001163999
NM_005241
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr18_-_46469118
|
0.837
|
NM_001190823
|
SMAD7
|
SMAD family member 7
|
|
chr2_+_203499819
|
0.835
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr9_+_115513050
|
0.833
|
NM_001012994
|
SNX30
|
sorting nexin family member 30
|
|
chr9_+_4490193
|
0.825
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr3_+_43328001
|
0.771
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr9_-_80646150
|
0.738
|
NM_002072
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide
|
|
chr7_-_86595203
|
0.737
|
NM_152748
|
KIAA1324L
|
KIAA1324-like
|
|
chr12_-_96184510
|
0.722
|
NM_021229
|
NTN4
|
netrin 4
|
|
chr11_-_89322778
|
0.701
|
NM_001143837
|
NOX4
|
NADPH oxidase 4
|
|
chr11_-_2906960
|
0.692
|
NM_000076
NM_001122630
NM_001122631
|
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2)
|
|
chr2_-_44588622
|
0.685
|
NM_001171606
NM_001171613
|
PREPL
|
prolyl endopeptidase-like
|
|
chr9_-_111882209
|
0.667
|
NM_032012
|
C9orf5
|
chromosome 9 open reading frame 5
|
|
chr2_+_109150807
|
0.665
|
NM_001193483
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr16_+_14164879
|
0.664
|
NM_014048
|
MKL2
|
MKL/myocardin-like 2
|
|
chr2_+_109237679
|
0.664
|
NM_001193484
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr2_+_46524448
|
0.662
|
NM_001430
|
EPAS1
|
endothelial PAS domain protein 1
|
|
chr11_-_89224413
|
0.647
|
NM_001143836
NM_016931
|
NOX4
|
NADPH oxidase 4
|
|
chr17_+_47865980
|
0.637
|
NM_001199155
NM_001199156
NM_001199157
NM_001199158
NM_007067
|
KAT7
|
K(lysine) acetyltransferase 7
|
|
chr2_+_235860616
|
0.637
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr12_-_56727720
|
0.627
|
NM_001166279
NM_014871
|
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr5_-_90679115
|
0.622
|
NM_020801
|
ARRDC3
|
arrestin domain containing 3
|
|
chrX_+_77166178
|
0.622
|
NM_000052
|
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide
|
|
chr12_+_3186520
|
0.618
|
NM_001168320
NM_006675
|
TSPAN9
|
tetraspanin 9
|
|
chr13_-_107187312
|
0.617
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr6_+_11538486
|
0.609
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr7_-_86688944
|
0.602
|
NM_001142749
|
KIAA1324L
|
KIAA1324-like
|
|
chr13_+_39612440
|
0.601
|
NM_001012754
NM_001017370
|
NHLRC3
|
NHL repeat containing 3
|
|
chr2_-_44587812
|
0.599
|
NM_006036
|
PREPL
|
prolyl endopeptidase-like
|
|
chr8_-_52811707
|
0.594
|
NM_052937
|
PCMTD1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1
|
|
chr8_-_81787015
|
0.590
|
NM_001033723
|
ZNF704
|
zinc finger protein 704
|
|
chr2_+_45878912
|
0.585
|
NM_005400
|
PRKCE
|
protein kinase C, epsilon
|
|
chr2_+_109204745
|
0.582
|
NM_004987
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr2_-_44588943
|
0.581
|
NM_001171603
NM_001171617
|
PREPL
|
prolyl endopeptidase-like
|
|
chr2_+_109271267
|
0.577
|
NM_001193485
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chrX_-_154033729
|
0.576
|
NM_001166460
NM_001166461
NM_001166462
NM_002436
|
MPP1
|
membrane protein, palmitoylated 1, 55kDa
|
|
chr12_-_56727446
|
0.572
|
NM_001127460
|
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr2_-_44586854
|
0.570
|
NM_001042385
NM_001042386
|
PREPL
|
prolyl endopeptidase-like
|
|
chr4_-_42658883
|
0.568
|
NM_001105529
NM_006095
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr7_+_89840999
|
0.563
|
NM_001244944
NM_001244945
NM_152999
NM_001040665
NM_001244946
|
STEAP2
|
STEAP family member 2, metalloreductase
|
|
chr15_-_56035176
|
0.560
|
NM_173814
|
PRTG
|
protogenin
|
|
chr2_+_109223600
|
0.559
|
NM_001193482
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr3_-_32023237
|
0.557
|
NM_001174060
NM_017784
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr14_+_24867926
|
0.557
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr3_+_194406614
|
0.555
|
NM_153690
|
FAM43A
|
family with sequence similarity 43, member A
|
|
chr1_+_25870043
|
0.549
|
NM_015627
|
LDLRAP1
|
low density lipoprotein receptor adaptor protein 1
|
|
chr14_-_54955714
|
0.543
|
NM_004124
|
GMFB
|
glia maturation factor, beta
|
|
chr4_+_140586921
|
0.538
|
NM_001204366
NM_001204367
NM_001204368
NM_002413
|
MGST2
|
microsomal glutathione S-transferase 2
|
|
chr17_+_34136437
|
0.534
|
NM_003487
NM_139215
|
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa
|
|
chr6_+_136172802
|
0.534
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr14_-_89883306
|
0.532
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr5_-_107006585
|
0.531
|
NM_001962
|
EFNA5
|
ephrin-A5
|
|
chr11_+_111848008
|
0.522
|
NM_033425
|
DIXDC1
|
DIX domain containing 1
|
|
chr1_+_178694273
|
0.515
|
NM_152663
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr1_-_94703252
|
0.514
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr3_-_169381417
|
0.511
|
NM_001205194
NM_004991
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr19_+_10736167
|
0.508
|
NM_020428
|
SLC44A2
|
solute carrier family 44, member 2
|
|
chr9_+_91003238
|
0.504
|
NM_006717
|
SPIN1
|
spindlin 1
|
|
chr19_+_10713055
|
0.499
|
NM_001145056
|
SLC44A2
|
solute carrier family 44, member 2
|
|
chr19_-_14316980
|
0.493
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr1_-_76076762
|
0.491
|
NM_001130058
NM_152697
|
SLC44A5
|
solute carrier family 44, member 5
|
|
chr2_+_109204904
|
0.489
|
NM_001193488
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr14_+_58765209
|
0.483
|
NM_002892
NM_023000
NM_023001
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr1_+_76540374
|
0.476
|
NM_001160011
NM_152996
|
ST6GALNAC3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3
|
|
chr11_-_27681195
|
0.466
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr6_-_41909386
|
0.459
|
NM_001136125
NM_001760
|
CCND3
|
cyclin D3
|
|
chr14_-_90085325
|
0.458
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr19_+_32897021
|
0.457
|
NM_207325
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr19_+_39897457
|
0.455
|
NM_003407
|
ZFP36
|
zinc finger protein 36, C3H type, homolog (mouse)
|
|
chr3_-_27763784
|
0.445
|
NM_005442
|
EOMES
|
eomesodermin
|
|
chr2_-_55646946
|
0.436
|
NM_001135597
NM_018084
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr19_+_32896515
|
0.436
|
NM_001172774
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr10_-_116286661
|
0.434
|
NM_006720
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr8_+_97505876
|
0.429
|
NM_002998
|
SDC2
|
syndecan 2
|
|
chr6_-_42016609
|
0.429
|
NM_001136017
NM_001136126
|
CCND3
|
cyclin D3
|
|
chr6_-_79787939
|
0.427
|
NM_017934
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr12_+_56521953
|
0.420
|
NM_001184796
NM_015292
|
ESYT1
|
extended synaptotagmin-like protein 1
|
|
chr10_-_116418056
|
0.420
|
NM_002313
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr10_+_129705299
|
0.418
|
NM_006504
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr14_+_66974124
|
0.412
|
NM_001024218
NM_020806
|
GPHN
|
gephyrin
|
|
chr10_-_116444374
|
0.410
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr11_+_92085261
|
0.404
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr3_+_23987514
|
0.401
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr12_-_31743822
|
0.400
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr7_+_30174343
|
0.398
|
NM_152793
|
C7orf41
|
chromosome 7 open reading frame 41
|
|
chr1_+_198125890
|
0.397
|
NM_133494
|
NEK7
|
NIMA (never in mitosis gene a)-related kinase 7
|
|
chr2_-_218808705
|
0.396
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr21_-_39870303
|
0.389
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr6_-_16761600
|
0.389
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr1_+_212208918
|
0.387
|
NM_016448
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr9_+_2621740
|
0.381
|
NM_001018056
NM_003383
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr8_-_95961543
|
0.381
|
NM_001135733
NM_033285
|
TP53INP1
|
tumor protein p53 inducible nuclear protein 1
|
|
chr9_-_16870719
|
0.379
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr1_-_236228476
|
0.378
|
NM_002508
|
NID1
|
nidogen 1
|
|
chr17_-_80656461
|
0.377
|
NM_006822
|
RAB40B
|
RAB40B, member RAS oncogene family
|
|
chr4_-_89744154
|
0.375
|
NM_001015045
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr5_+_86564044
|
0.375
|
NM_002890
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr21_-_40033617
|
0.373
|
NM_001243432
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr3_-_66024508
|
0.371
|
NM_001033057
NM_004742
NM_015520
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1
|
|
chr5_+_86564727
|
0.367
|
NM_022650
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr11_-_27721975
|
0.367
|
NM_001143813
NM_001143814
NM_001709
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr10_+_35535952
|
0.364
|
NM_181698
|
CCNY
|
cyclin Y
|
|
chr18_-_72921280
|
0.363
|
NM_175907
|
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2
|
|
chr14_+_63671101
|
0.356
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr8_-_93978264
|
0.356
|
NM_001191035
NM_001171795
NM_001171796
NM_001171797
NM_001171798
NM_001171799
|
C8orf83
|
chromosome 8 open reading frame 83
|
|
chr1_+_84764048
|
0.355
|
NM_001010971
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr1_+_84768333
|
0.354
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr1_+_84767288
|
0.353
|
NM_001134663
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr11_-_27722446
|
0.347
|
NM_001143812
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr6_+_150070818
|
0.346
|
NM_001252049
NM_001252050
NM_001252051
NM_001252052
NM_001252053
NM_005389
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase
|
|
chrX_-_114468604
|
0.346
|
NM_001243963
NM_020871
|
LRCH2
|
leucine-rich repeats and calponin homology (CH) domain containing 2
|
|
chr2_-_158731622
|
0.345
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr9_+_36036900
|
0.343
|
NM_021111
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs
|
|
chr11_-_27722599
|
0.343
|
NM_001143808
NM_001143809
NM_001143810
NM_001143811
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27721179
|
0.340
|
NM_170734
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr1_+_84609941
|
0.340
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr8_-_93977715
|
0.339
|
NM_001191036
|
C8orf83
|
chromosome 8 open reading frame 83
|
|
chr10_+_129845806
|
0.339
|
NM_130435
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr11_-_27742224
|
0.338
|
NM_001143805
NM_001143806
NM_170732
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr6_+_17600504
|
0.337
|
NM_016255
|
FAM8A1
|
family with sequence similarity 8, member A1
|
|
chr1_+_84630575
|
0.336
|
NM_001242860
NM_001242861
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr8_-_9008138
|
0.335
|
NM_024607
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B
|
|
chr1_+_84630014
|
0.333
|
NM_001242857
NM_001242858
NM_001242859
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr6_-_32157511
|
0.333
|
NM_002586
|
PBX2
|
pre-B-cell leukemia homeobox 2
|
|
chrX_-_34675366
|
0.327
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr17_+_61699786
|
0.325
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr1_-_109935869
|
0.324
|
NM_001205228
|
SORT1
|
sortilin 1
|
|
chr11_-_27723061
|
0.320
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr9_-_79520878
|
0.319
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr1_+_164528586
|
0.313
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr10_-_101945787
|
0.311
|
NM_001100626
|
ERLIN1
|
ER lipid raft associated 1
|
|
chr6_+_88182624
|
0.311
|
NM_001168398
NM_006416
|
SLC35A1
|
solute carrier family 35 (CMP-sialic acid transporter), member A1
|
|
chr3_+_135684463
|
0.309
|
NM_001190447
NM_002718
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr11_-_27743604
|
0.309
|
NM_170731
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr10_-_101945686
|
0.308
|
NM_006459
|
ERLIN1
|
ER lipid raft associated 1
|
|
chr5_+_118407067
|
0.308
|
NM_005509
|
DMXL1
|
Dmx-like 1
|
|
chr1_+_84647342
|
0.306
|
NM_001242862
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr7_+_142985307
|
0.306
|
NM_032982
NM_032983
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase
|
|
chr3_-_64211111
|
0.306
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr10_+_35625608
|
0.305
|
NM_145012
|
CCNY
|
cyclin Y
|
|
chr2_-_62733475
|
0.303
|
NM_198276
|
TMEM17
|
transmembrane protein 17
|
|
chr11_-_27741192
|
0.303
|
NM_001143807
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr3_+_135741576
|
0.302
|
NM_181897
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr21_-_44846927
|
0.300
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr22_-_36903086
|
0.300
|
NM_024955
NM_001102371
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2
|
|
chr10_-_65225605
|
0.299
|
NM_032776
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr8_-_9009050
|
0.299
|
NM_001201329
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B
|
|
chr2_+_61292999
|
0.293
|
NM_001129993
|
KIAA1841
|
KIAA1841
|
|
chr1_+_33004771
|
0.289
|
NM_001040441
|
ZBTB8A
|
zinc finger and BTB domain containing 8A
|
|
chr8_-_103137134
|
0.289
|
NM_001040630
|
NCALD
|
neurocalcin delta
|
|
chr1_-_92371558
|
0.288
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr2_+_241375030
|
0.287
|
NM_002081
|
GPC1
|
glypican 1
|
|
chr7_-_112579726
|
0.281
|
NM_152556
|
C7orf60
|
chromosome 7 open reading frame 60
|
|
chr8_-_102803438
|
0.280
|
NM_032041
|
NCALD
|
neurocalcin delta
|
|
chr16_+_69220940
|
0.278
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr22_-_39548448
|
0.277
|
NM_175709
|
CBX7
|
chromobox homolog 7
|
|
chr1_-_92351613
|
0.274
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr1_-_8483701
|
0.272
|
NM_001042682
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats
|
|
chr18_-_812268
|
0.271
|
NM_005433
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1
|
|
chr10_-_62703822
|
0.271
|
NM_001242359
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr3_+_23986735
|
0.271
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr1_+_201798281
|
0.270
|
NM_018085
|
IPO9
|
importin 9
|
|
chr2_+_121010413
|
0.270
|
NM_002881
|
RALB
|
v-ral simian leukemia viral oncogene homolog B (ras related; GTP binding protein)
|
|
chr20_-_60640842
|
0.269
|
NM_003185
|
TAF4
|
TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa
|
|
chr11_+_7535000
|
0.268
|
NM_003621
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|