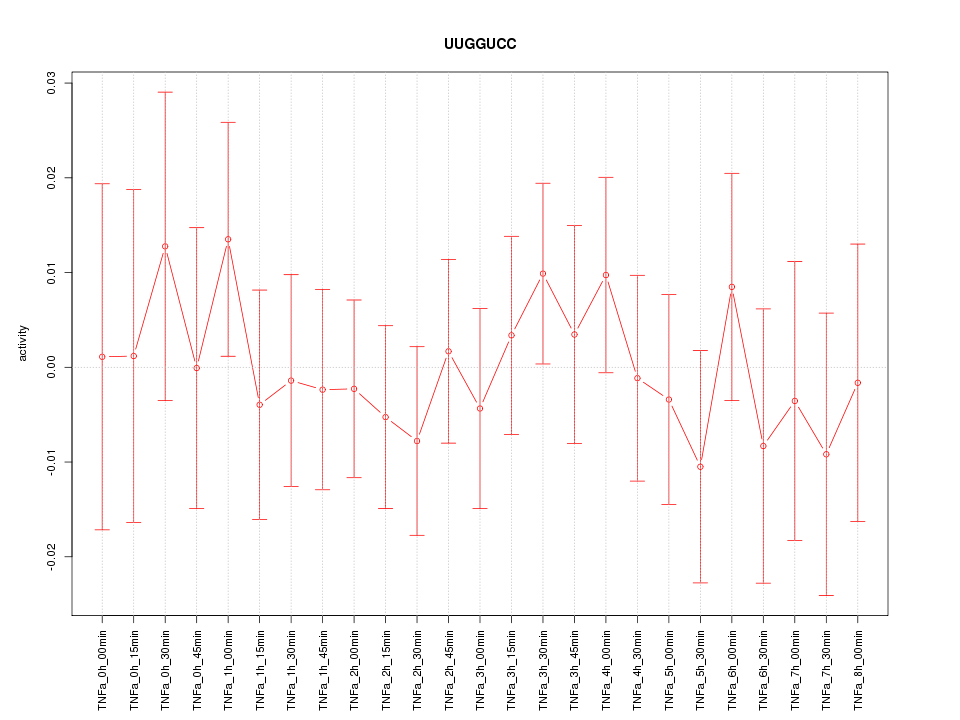
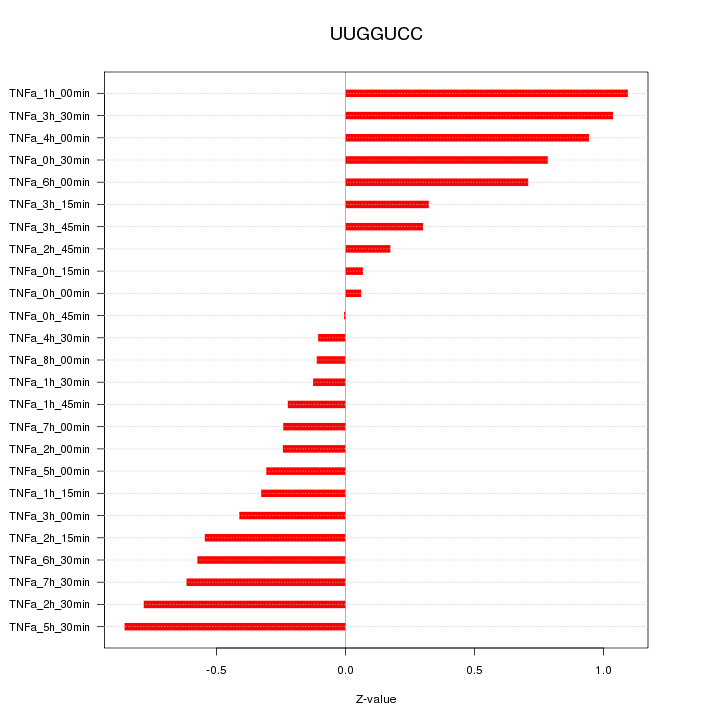
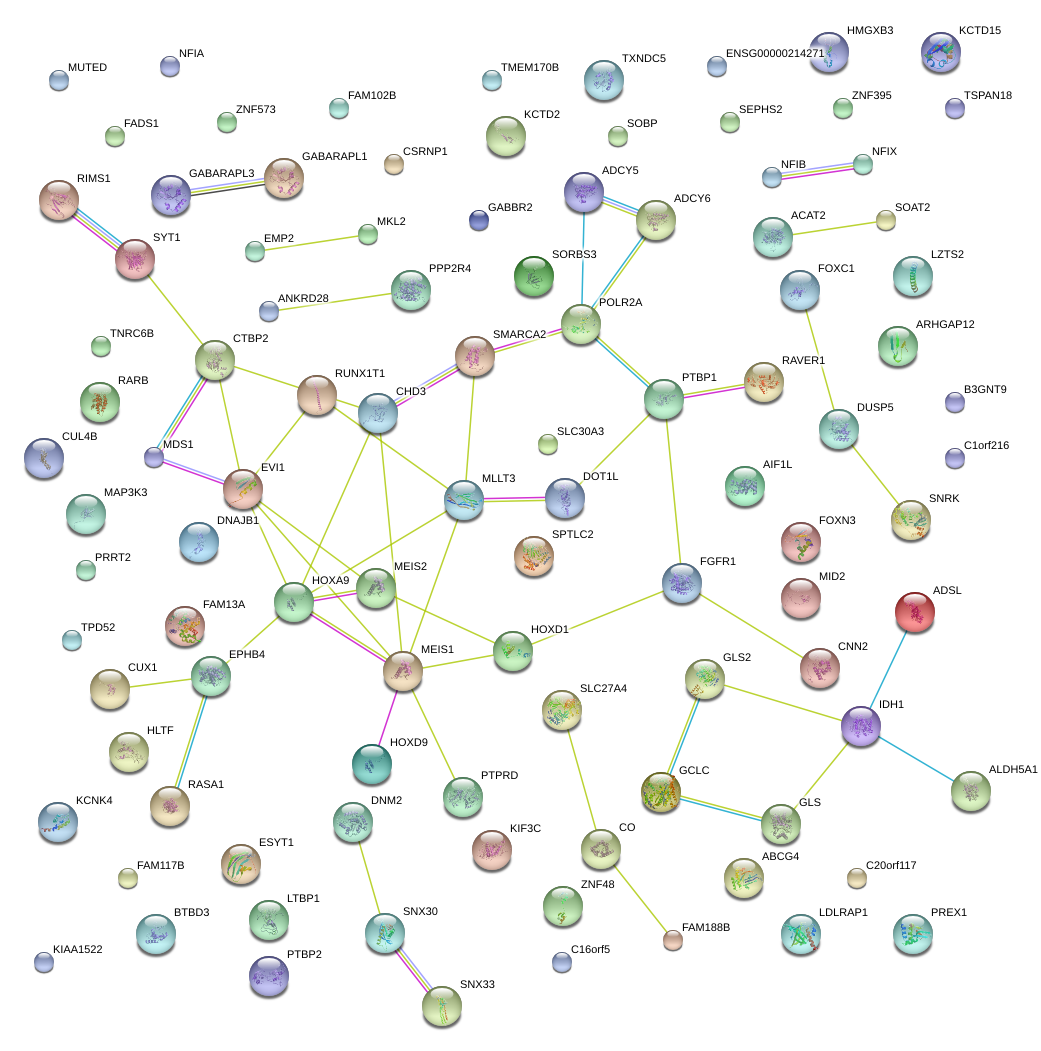
|
chr8_-_93029864
|
0.807
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93111915
|
0.775
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93107881
|
0.766
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93107442
|
0.728
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr7_+_30960914
|
0.711
|
NM_001185060
NM_001185061
NM_001185062
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr8_-_93115453
|
0.693
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93075190
|
0.679
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr9_+_133971862
|
0.629
|
NM_001185095
NM_001185096
NM_031426
|
AIF1L
|
allograft inflammatory factor 1-like
|
|
chr7_+_30951414
|
0.623
|
NM_198098
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr3_+_43328001
|
0.594
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr1_+_33231227
|
0.577
|
NM_001198972
NM_001198973
|
KIAA1522
|
KIAA1522
|
|
chr1_+_61542928
|
0.576
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_61547625
|
0.573
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_25870043
|
0.569
|
NM_015627
|
LDLRAP1
|
low density lipoprotein receptor adaptor protein 1
|
|
chr1_+_61547388
|
0.567
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr8_-_28243941
|
0.562
|
NM_018660
|
ZNF395
|
zinc finger protein 395
|
|
chr1_+_33207343
|
0.542
|
NM_020888
|
KIAA1522
|
KIAA1522
|
|
chr9_-_8733893
|
0.519
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr9_-_10612722
|
0.517
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr10_+_112257624
|
0.483
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr6_+_160182988
|
0.475
|
NM_005891
|
ACAT2
|
acetyl-CoA acetyltransferase 2
|
|
chr9_-_14398981
|
0.473
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr7_-_100425035
|
0.451
|
NM_004444
|
EPHB4
|
EPH receptor B4
|
|
chr20_-_47444404
|
0.447
|
NM_020820
|
PREX1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1
|
|
chr2_+_177053306
|
0.434
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr16_+_14164879
|
0.434
|
NM_014048
|
MKL2
|
MKL/myocardin-like 2
|
|
chr9_-_14314036
|
0.420
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr1_+_109102970
|
0.414
|
NM_001010883
|
FAM102B
|
family with sequence similarity 102, member B
|
|
chr6_+_1610680
|
0.402
|
NM_001453
|
FOXC1
|
forkhead box C1
|
|
chr2_+_203499819
|
0.397
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr2_-_209119751
|
0.389
|
NM_005896
|
IDH1
|
isocitrate dehydrogenase 1 (NADP+), soluble
|
|
chr3_-_168865521
|
0.388
|
NM_001164000
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr2_+_66662516
|
0.387
|
NM_002398
|
MEIS1
|
Meis homeobox 1
|
|
chr3_-_15900928
|
0.384
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr3_-_168864325
|
0.377
|
NM_001105078
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr3_-_168864066
|
0.375
|
NM_001105077
NM_001163999
NM_005241
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr3_-_15839674
|
0.368
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr6_+_11538486
|
0.363
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr6_+_24495166
|
0.361
|
NM_001080
NM_170740
|
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1
|
|
chrX_+_107069081
|
0.340
|
NM_012216
NM_052817
|
MID2
|
midline 2
|
|
chr8_-_80993009
|
0.335
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr8_-_38326351
|
0.329
|
NM_001174063
NM_001174064
NM_015850
NM_023105
NM_023106
NM_023110
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr11_+_119019749
|
0.328
|
NM_022169
|
ABCG4
|
ATP-binding cassette, sub-family G (WHITE), member 4
|
|
chr3_-_39195101
|
0.322
|
NM_033027
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1
|
|
chr16_+_29823408
|
0.322
|
NM_145239
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr20_+_11898536
|
0.314
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr11_+_119020298
|
0.313
|
NM_001142505
|
ABCG4
|
ATP-binding cassette, sub-family G (WHITE), member 4
|
|
chr11_+_64058772
|
0.312
|
NM_033310
|
KCNK4
|
potassium channel, subfamily K, member 4
|
|
chr20_+_11871364
|
0.310
|
NM_181443
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr6_+_107811272
|
0.307
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr19_-_14629160
|
0.306
|
NM_006145
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1
|
|
chr9_-_20622476
|
0.301
|
NM_004529
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3
|
|
chr16_-_10674443
|
0.300
|
NM_001424
|
EMP2
|
epithelial membrane protein 2
|
|
chr12_+_10365415
|
0.297
|
NM_031412
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1
|
|
chr10_-_126716452
|
0.295
|
NM_022802
|
CTBP2
|
C-terminal binding protein 2
|
|
chr8_-_81083660
|
0.283
|
NM_001025253
NM_005079
|
TPD52
|
tumor protein D52
|
|
chr11_+_44785975
|
0.283
|
NM_130783
|
TSPAN18
|
tetraspanin 18
|
|
chr17_+_61699786
|
0.280
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr2_+_33359607
|
0.279
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr2_+_33172368
|
0.279
|
NM_206943
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr8_-_38325241
|
0.278
|
NM_001174065
NM_001174066
NM_001174067
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr10_+_102756829
|
0.271
|
NM_032429
|
LZTS2
|
leucine zipper, putative tumor suppressor 2
|
|
chr9_+_115513050
|
0.271
|
NM_001012994
|
SNX30
|
sorting nexin family member 30
|
|
chr19_-_10444186
|
0.270
|
NM_133452
|
RAVER1
|
ribonucleoprotein, PTB-binding 1
|
|
chr6_-_53409897
|
0.266
|
NM_001197115
NM_001498
|
GCLC
|
glutamate-cysteine ligase, catalytic subunit
|
|
chr14_-_78083109
|
0.265
|
NM_004863
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2
|
|
chr10_-_126849066
|
0.263
|
NM_001329
|
CTBP2
|
C-terminal binding protein 2
|
|
chr2_-_27485774
|
0.262
|
NM_003459
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3
|
|
chr2_+_191745546
|
0.251
|
NM_014905
|
GLS
|
glutaminase
|
|
chr10_-_126849556
|
0.249
|
NM_001083914
|
CTBP2
|
C-terminal binding protein 2
|
|
chr10_-_32217762
|
0.248
|
NM_018287
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chr16_-_67184822
|
0.247
|
NM_033309
|
B3GNT9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9
|
|
chr2_-_26205437
|
0.247
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chr3_-_15798057
|
0.246
|
NM_001195099
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr7_+_101460881
|
0.244
|
NM_181552
|
CUX1
|
cut-like homeobox 1
|
|
chr17_+_7387688
|
0.244
|
NM_000937
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa
|
|
chr1_+_97187174
|
0.242
|
NM_021190
|
PTBP2
|
polypyrimidine tract binding protein 2
|
|
chr20_-_35488275
|
0.240
|
NM_199181
|
KIAA0889
|
KIAA0889
|
|
chr17_+_73043154
|
0.238
|
NM_015353
|
KCTD2
|
potassium channel tetramerisation domain containing 2
|
|
chr1_-_36184727
|
0.238
|
NM_152374
|
C1orf216
|
chromosome 1 open reading frame 216
|
|
chr6_+_72922513
|
0.235
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr4_-_89744154
|
0.223
|
NM_001015045
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr19_+_34287662
|
0.221
|
NM_001129994
NM_001129995
NM_024076
|
KCTD15
|
potassium channel tetramerisation domain containing 15
|
|
chr14_-_90085325
|
0.215
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr19_+_1026271
|
0.214
|
NM_004368
NM_201277
|
CNN2
|
calponin 2
|
|
chr9_+_2015218
|
0.213
|
NM_003070
NM_139045
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2
|
|
chr6_-_7910278
|
0.213
|
NM_001145549
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum)
|
|
chr8_+_22423170
|
0.212
|
NM_001018003
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr6_+_73076431
|
0.210
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr6_+_72926144
|
0.209
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chrX_-_119709618
|
0.207
|
NM_003588
|
CUL4B
|
cullin 4B
|
|
chr5_+_149380168
|
0.206
|
NM_014983
|
HMGXB3
|
HMG box domain containing 3
|
|
chr16_+_30406432
|
0.204
|
NM_001214909
|
ZNF48
|
zinc finger protein 48
|
|
chr3_-_169381417
|
0.203
|
NM_001205194
NM_004991
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr19_+_797390
|
0.203
|
NM_002819
NM_031990
NM_031991
NM_175847
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chr3_-_148804122
|
0.202
|
NM_003071
NM_139048
|
HLTF
|
helicase-like transcription factor
|
|
chr22_+_40573879
|
0.198
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr16_+_30389632
|
0.198
|
NM_001214906
NM_001214907
|
ZNF48
|
zinc finger protein 48
|
|
chr12_+_79257772
|
0.196
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr9_-_101470836
|
0.195
|
NM_005458
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2
|
|
chr19_+_2164119
|
0.194
|
NM_032482
|
DOT1L
|
DOT1-like, histone H3 methyltransferase (S. cerevisiae)
|
|
chr3_-_123135188
|
0.192
|
NM_001199642
|
ADCY5
|
adenylate cyclase 5
|
|
chr9_+_131102786
|
0.189
|
NM_005094
|
SLC27A4
|
solute carrier family 27 (fatty acid transporter), member 4
|
|
chr9_+_131873592
|
0.189
|
NM_178000
NM_178001
NM_178003
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4
|
|
chr16_-_30457197
|
0.188
|
NM_012248
|
SEPHS2
|
selenophosphate synthetase 2
|
|
chr17_+_7788094
|
0.188
|
NM_001005271
|
CHD3
|
chromodomain helicase DNA binding protein 3
|
|
chr12_+_79439432
|
0.188
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr12_+_56521953
|
0.187
|
NM_001184796
NM_015292
|
ESYT1
|
extended synaptotagmin-like protein 1
|
|
chr14_-_89883306
|
0.186
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr11_-_61584451
|
0.184
|
NM_013402
|
FADS1
|
fatty acid desaturase 1
|
|
chr16_+_30406711
|
0.184
|
NM_152652
|
ZNF48
|
zinc finger protein 48
|
|
chr3_+_25469833
|
0.182
|
NM_000965
|
RARB
|
retinoic acid receptor, beta
|
|
chr16_-_4588379
|
0.181
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr6_+_72926462
|
0.181
|
NM_001168410
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_-_29450406
|
0.181
|
NM_001003682
|
TMEM200B
|
transmembrane protein 200B
|
|
chr7_-_27205148
|
0.178
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr9_+_131873752
|
0.178
|
NM_001193397
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4
|
|
chr13_-_110438896
|
0.174
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr15_-_37393364
|
0.174
|
NM_001220482
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr8_-_127570710
|
0.174
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr6_-_80656870
|
0.173
|
NM_022726
|
ELOVL4
|
ELOVL fatty acid elongase 4
|
|
chr20_-_3388219
|
0.171
|
NM_001009984
|
C20orf194
|
chromosome 20 open reading frame 194
|
|
chr5_-_172198160
|
0.170
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr17_-_47841517
|
0.169
|
NM_030802
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr7_-_138665853
|
0.168
|
NM_001164665
NM_020910
|
KIAA1549
|
KIAA1549
|
|
chr1_-_29449012
|
0.168
|
NM_001171868
|
TMEM200B
|
transmembrane protein 200B
|
|
chrX_-_119694719
|
0.167
|
NM_001079872
|
CUL4B
|
cullin 4B
|
|
chr2_-_69870976
|
0.165
|
NM_014911
|
AAK1
|
AP2 associated kinase 1
|
|
chrX_-_25034064
|
0.162
|
NM_139058
|
ARX
|
aristaless related homeobox
|
|
chr6_-_119031230
|
0.162
|
NM_001178035
|
CEP85L
|
centrosomal protein 85kDa-like
|
|
chr8_-_66753968
|
0.162
|
NM_001242318
|
PDE7A
|
phosphodiesterase 7A
|
|
chr15_-_37390372
|
0.161
|
NM_172315
|
MEIS2
|
Meis homeobox 2
|
|
chr16_-_4588738
|
0.159
|
NM_001199055
NM_001199056
NM_013399
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr8_-_145838887
|
0.158
|
NM_025251
|
ARHGAP39
|
Rho GTPase activating protein 39
|
|
chr15_+_91411884
|
0.156
|
NM_002569
|
FURIN
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr8_+_22409226
|
0.156
|
NM_005775
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr14_+_24899139
|
0.153
|
NM_015299
|
KHNYN
|
KH and NYN domain containing
|
|
chr17_-_39942937
|
0.150
|
NM_002230
NM_021991
|
JUP
|
junction plakoglobin
|
|
chr17_+_60704747
|
0.150
|
NM_006039
|
MRC2
|
mannose receptor, C type 2
|
|
chr3_-_196159304
|
0.148
|
NM_015562
|
UBXN7
|
UBX domain protein 7
|
|
chr17_-_27045172
|
0.147
|
NM_001142624
NM_001142625
NM_001144943
|
RAB34
|
RAB34, member RAS oncogene family
|
|
chr3_-_186524226
|
0.146
|
NM_002916
|
RFC4
|
replication factor C (activator 1) 4, 37kDa
|
|
chr3_-_186524483
|
0.146
|
NM_181573
|
RFC4
|
replication factor C (activator 1) 4, 37kDa
|
|
chr6_-_7910716
|
0.145
|
NM_030810
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum)
|
|
chr6_+_21593963
|
0.145
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr5_+_43121641
|
0.143
|
NM_003432
|
ZNF131
|
zinc finger protein 131
|
|
chr12_+_79258448
|
0.143
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr8_+_38034066
|
0.142
|
NM_001204878
NM_004874
|
BAG4
|
BCL2-associated athanogene 4
|
|
chr22_+_40440664
|
0.142
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr13_+_58205788
|
0.141
|
NM_001040429
|
PCDH17
|
protocadherin 17
|
|
chr19_-_7293876
|
0.141
|
NM_000208
NM_001079817
|
INSR
|
insulin receptor
|
|
chr8_-_23021447
|
0.141
|
NM_003840
|
TNFRSF10D
|
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain
|
|
chr17_-_79881304
|
0.140
|
NM_032711
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr9_+_131873227
|
0.138
|
NM_021131
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4
|
|
chr3_-_123167391
|
0.138
|
NM_183357
|
ADCY5
|
adenylate cyclase 5
|
|
chr11_-_88781039
|
0.133
|
NM_001143831
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chrX_-_140271293
|
0.132
|
NM_012317
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr11_-_118927872
|
0.132
|
NM_006389
NM_001130991
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr17_-_48227873
|
0.132
|
NM_032595
|
PPP1R9B
|
protein phosphatase 1, regulatory subunit 9B
|
|
chr19_-_14316980
|
0.132
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr2_-_130939146
|
0.130
|
NM_001171083
NM_017751
NM_017951
|
SMPD4
|
sphingomyelin phosphodiesterase 4, neutral membrane (neutral sphingomyelinase-3)
|
|
chr15_-_37391596
|
0.130
|
NM_172316
|
MEIS2
|
Meis homeobox 2
|
|
chr12_-_50222144
|
0.129
|
NM_001037806
|
NCKAP5L
|
NCK-associated protein 5-like
|
|
chr12_+_56137063
|
0.129
|
NM_005811
|
GDF11
|
growth differentiation factor 11
|
|
chr12_+_15699277
|
0.128
|
NM_030668
NM_030669
NM_030670
NM_030671
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr3_+_5229357
|
0.126
|
NM_014674
|
EDEM1
|
ER degradation enhancer, mannosidase alpha-like 1
|
|
chr8_+_29952921
|
0.126
|
NM_001128208
NM_015344
|
LEPROTL1
|
leptin receptor overlapping transcript-like 1
|
|
chr4_-_141075232
|
0.126
|
NM_018717
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr12_-_82153092
|
0.126
|
NM_003625
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr3_-_13114547
|
0.125
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr7_+_102073953
|
0.122
|
NM_001126340
NM_032831
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr13_+_98795410
|
0.122
|
NM_001001715
NM_005766
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr16_-_66584045
|
0.122
|
NM_001172643
NM_001172644
NM_001172645
NM_004614
|
TK2
|
thymidine kinase 2, mitochondrial
|
|
chr18_-_812268
|
0.122
|
NM_005433
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1
|
|
chr6_-_166075520
|
0.120
|
NM_006661
NM_001130690
|
PDE10A
|
phosphodiesterase 10A
|
|
chr17_+_7792034
|
0.120
|
NM_001005273
NM_005852
|
CHD3
|
chromodomain helicase DNA binding protein 3
|
|
chr17_+_635656
|
0.120
|
NM_024792
|
FAM57A
|
family with sequence similarity 57, member A
|
|
chr17_-_7387513
|
0.119
|
NM_020899
|
ZBTB4
|
zinc finger and BTB domain containing 4
|
|
chr1_+_11866206
|
0.117
|
NM_001286
NM_021735
NM_021736
NM_021737
|
CLCN6
|
chloride channel 6
|
|
chr17_-_66287256
|
0.117
|
NM_001174166
NM_004694
|
SLC16A6
|
solute carrier family 16, member 6 (monocarboxylic acid transporter 7)
|
|
chr1_-_179112178
|
0.116
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr12_+_50478942
|
0.114
|
NM_003076
NM_139071
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1
|
|
chr16_-_1429612
|
0.114
|
NM_001193389
NM_023076
|
UNKL
|
unkempt homolog (Drosophila)-like
|
|
chr19_-_46272496
|
0.113
|
NM_175875
|
SIX5
|
SIX homeobox 5
|
|
chr16_+_58549382
|
0.112
|
NM_001160305
NM_024860
|
SETD6
|
SET domain containing 6
|
|
chr17_-_7382943
|
0.110
|
NM_001128833
|
ZBTB4
|
zinc finger and BTB domain containing 4
|
|
chr17_-_79885544
|
0.109
|
NM_002359
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr8_+_67039130
|
0.106
|
NM_033058
NM_184085
NM_184086
NM_184087
|
TRIM55
|
tripartite motif containing 55
|
|
chr17_-_53800074
|
0.106
|
NM_018286
|
TMEM100
|
transmembrane protein 100
|
|
chr12_-_54778456
|
0.104
|
NM_015481
|
ZNF385A
|
zinc finger protein 385A
|
|
chr15_+_85359910
|
0.103
|
NM_020778
|
ALPK3
|
alpha-kinase 3
|
|
chr1_+_32573619
|
0.103
|
NM_012316
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7)
|
|
chr4_+_111397181
|
0.102
|
NM_001977
|
ENPEP
|
glutamyl aminopeptidase (aminopeptidase A)
|
|
chr17_-_53809444
|
0.102
|
NM_001099640
|
TMEM100
|
transmembrane protein 100
|
|
chr17_+_80477564
|
0.101
|
NM_004514
|
FOXK2
|
forkhead box K2
|
|
chr17_-_27044728
|
0.101
|
NM_001144942
NM_031934
|
RAB34
|
RAB34, member RAS oncogene family
|
|
chr15_-_37392358
|
0.101
|
NM_170674
NM_170675
NM_170676
NM_170677
|
MEIS2
|
Meis homeobox 2
|
|
chr2_-_197791339
|
0.101
|
NM_024989
|
PGAP1
|
post-GPI attachment to proteins 1
|
|
chr16_-_15736964
|
0.101
|
NM_001184998
NM_001184999
NM_014647
|
KIAA0430
|
KIAA0430
|
|
chr9_-_37034358
|
0.100
|
NM_016734
|
PAX5
|
paired box 5
|