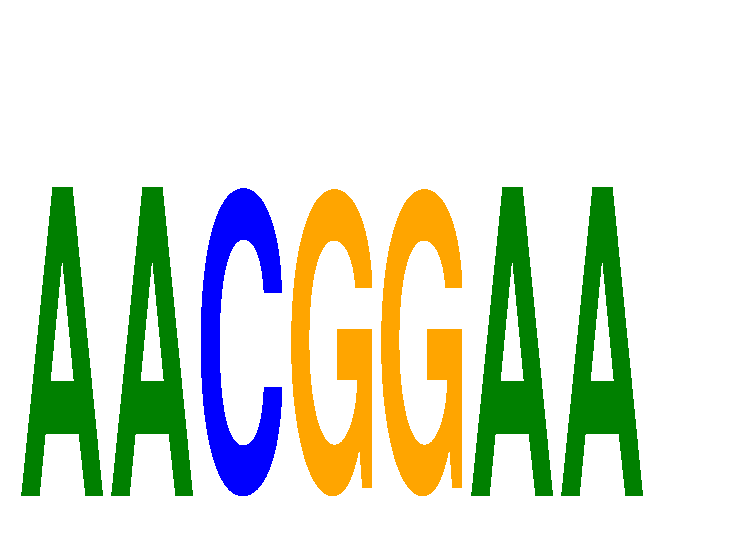Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for AACGGAA
Z-value: 0.08
miRNA associated with seed AACGGAA
| Name | miRBASE accession |
|---|---|
|
hsa-miR-191-5p
|
MIMAT0000440 |
Activity profile of AACGGAA motif
Sorted Z-values of AACGGAA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_208634287 | 0.04 |
ENST00000295417.3
|
FZD5
|
frizzled family receptor 5 |
| chr1_+_61547894 | 0.04 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr10_-_81205373 | 0.03 |
ENST00000372336.3
|
ZCCHC24
|
zinc finger, CCHC domain containing 24 |
| chr17_-_7232585 | 0.03 |
ENST00000571887.1
ENST00000315614.7 ENST00000399464.2 ENST00000570460.1 |
NEURL4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr22_+_51112800 | 0.02 |
ENST00000414786.2
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr7_-_92463210 | 0.02 |
ENST00000265734.4
|
CDK6
|
cyclin-dependent kinase 6 |
| chr1_-_19283163 | 0.01 |
ENST00000455833.2
|
IFFO2
|
intermediate filament family orphan 2 |
| chr17_-_46623441 | 0.01 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chrX_-_41782249 | 0.01 |
ENST00000442742.2
ENST00000421587.2 ENST00000378166.4 ENST00000318588.9 ENST00000361962.4 |
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr8_-_134115118 | 0.01 |
ENST00000395352.3
ENST00000338087.5 |
SLA
|
Src-like-adaptor |
| chr2_-_128643496 | 0.01 |
ENST00000272647.5
|
AMMECR1L
|
AMMECR1-like |
| chr11_+_69061594 | 0.01 |
ENST00000441339.2
ENST00000308946.3 ENST00000535407.1 |
MYEOV
|
myeloma overexpressed |
| chr1_-_17766198 | 0.01 |
ENST00000375436.4
|
RCC2
|
regulator of chromosome condensation 2 |
| chr14_+_36295504 | 0.01 |
ENST00000216807.7
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr10_+_114709999 | 0.01 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr9_+_133454943 | 0.01 |
ENST00000319725.9
|
FUBP3
|
far upstream element (FUSE) binding protein 3 |
| chr3_-_18466787 | 0.01 |
ENST00000338745.6
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr1_+_33722080 | 0.01 |
ENST00000483388.1
ENST00000539719.1 |
ZNF362
|
zinc finger protein 362 |
| chr7_-_71801980 | 0.00 |
ENST00000329008.5
|
CALN1
|
calneuron 1 |
| chr6_+_116601265 | 0.00 |
ENST00000452085.3
|
DSE
|
dermatan sulfate epimerase |
| chr8_-_141645645 | 0.00 |
ENST00000519980.1
ENST00000220592.5 |
AGO2
|
argonaute RISC catalytic component 2 |
| chr3_+_115342159 | 0.00 |
ENST00000305124.6
|
GAP43
|
growth associated protein 43 |
| chr9_-_16870704 | 0.00 |
ENST00000380672.4
ENST00000380667.2 ENST00000380666.2 ENST00000486514.1 |
BNC2
|
basonuclin 2 |
| chr5_+_149887672 | 0.00 |
ENST00000261797.6
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr18_+_32558208 | 0.00 |
ENST00000436190.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr6_+_21593972 | 0.00 |
ENST00000244745.1
ENST00000543472.1 |
SOX4
|
SRY (sex determining region Y)-box 4 |
| chr12_-_58240470 | 0.00 |
ENST00000548823.1
ENST00000398073.2 |
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AACGGAA
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |