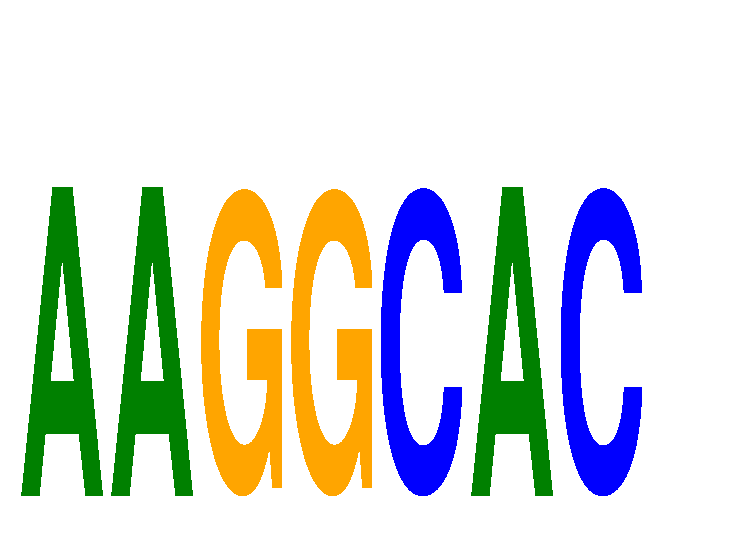Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for AAGGCAC
Z-value: 0.57
miRNA associated with seed AAGGCAC
| Name | miRBASE accession |
|---|---|
|
hsa-miR-124-3p.1
|
MIMAT0000422 |
Activity profile of AAGGCAC motif
Sorted Z-values of AAGGCAC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_72385437 | 0.59 |
ENST00000418754.2
ENST00000542969.2 ENST00000334456.5 |
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr2_+_112812778 | 0.57 |
ENST00000283206.4
|
TMEM87B
|
transmembrane protein 87B |
| chr5_-_59189545 | 0.54 |
ENST00000340635.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr3_-_164913777 | 0.48 |
ENST00000475390.1
|
SLITRK3
|
SLIT and NTRK-like family, member 3 |
| chr5_+_127419449 | 0.47 |
ENST00000262461.2
ENST00000343225.4 |
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr7_-_25019760 | 0.47 |
ENST00000352860.1
ENST00000353930.1 ENST00000431825.2 ENST00000313367.2 |
OSBPL3
|
oxysterol binding protein-like 3 |
| chr17_-_8151353 | 0.45 |
ENST00000315684.8
|
CTC1
|
CTS telomere maintenance complex component 1 |
| chr7_-_108096822 | 0.45 |
ENST00000379028.3
ENST00000413765.2 ENST00000379022.4 |
NRCAM
|
neuronal cell adhesion molecule |
| chr3_+_29322803 | 0.44 |
ENST00000396583.3
ENST00000383767.2 |
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr5_-_179780312 | 0.42 |
ENST00000253778.8
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr6_-_134639180 | 0.42 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr6_+_71998506 | 0.41 |
ENST00000370435.4
|
OGFRL1
|
opioid growth factor receptor-like 1 |
| chr20_-_18038521 | 0.41 |
ENST00000278780.6
|
OVOL2
|
ovo-like zinc finger 2 |
| chr1_-_169455169 | 0.40 |
ENST00000367804.4
ENST00000236137.5 |
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr4_-_80994210 | 0.40 |
ENST00000403729.2
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr13_-_44361025 | 0.39 |
ENST00000261488.6
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr14_+_21538429 | 0.37 |
ENST00000298694.4
ENST00000555038.1 |
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr15_+_79724858 | 0.36 |
ENST00000305428.3
|
KIAA1024
|
KIAA1024 |
| chr4_-_120550146 | 0.36 |
ENST00000354960.3
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific |
| chr4_+_95679072 | 0.36 |
ENST00000515059.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr1_+_65613217 | 0.35 |
ENST00000545314.1
|
AK4
|
adenylate kinase 4 |
| chr12_-_57472522 | 0.35 |
ENST00000379391.3
ENST00000300128.4 |
TMEM194A
|
transmembrane protein 194A |
| chr6_-_33756867 | 0.35 |
ENST00000293760.5
|
LEMD2
|
LEM domain containing 2 |
| chr1_-_197169672 | 0.34 |
ENST00000367405.4
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chr4_+_124320665 | 0.33 |
ENST00000394339.2
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr16_+_50187556 | 0.32 |
ENST00000561678.1
ENST00000357464.3 |
PAPD5
|
PAP associated domain containing 5 |
| chr3_-_185216766 | 0.32 |
ENST00000296254.3
|
TMEM41A
|
transmembrane protein 41A |
| chr8_+_23386305 | 0.30 |
ENST00000519973.1
|
SLC25A37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr21_+_27011584 | 0.30 |
ENST00000400532.1
ENST00000480456.1 ENST00000312957.5 |
JAM2
|
junctional adhesion molecule 2 |
| chr3_-_122283424 | 0.30 |
ENST00000477522.2
ENST00000360356.2 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr8_+_17354587 | 0.30 |
ENST00000494857.1
ENST00000522656.1 |
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr11_+_58939965 | 0.29 |
ENST00000227451.3
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr8_-_37756972 | 0.29 |
ENST00000330843.4
ENST00000522727.1 ENST00000287263.4 |
RAB11FIP1
|
RAB11 family interacting protein 1 (class I) |
| chr12_+_121570631 | 0.28 |
ENST00000546057.1
ENST00000377162.2 ENST00000328963.5 ENST00000535250.1 ENST00000541446.1 |
P2RX7
|
purinergic receptor P2X, ligand-gated ion channel, 7 |
| chr1_+_165796753 | 0.28 |
ENST00000367879.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr9_+_140500087 | 0.27 |
ENST00000371421.4
|
ARRDC1
|
arrestin domain containing 1 |
| chr6_-_154677900 | 0.27 |
ENST00000265198.4
ENST00000520261.1 |
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr6_+_7107999 | 0.27 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr1_+_168250194 | 0.27 |
ENST00000367821.3
|
TBX19
|
T-box 19 |
| chr1_+_78245303 | 0.26 |
ENST00000370791.3
ENST00000443751.2 |
FAM73A
|
family with sequence similarity 73, member A |
| chr10_-_105615164 | 0.26 |
ENST00000355946.2
ENST00000369774.4 |
SH3PXD2A
|
SH3 and PX domains 2A |
| chrX_+_68048803 | 0.25 |
ENST00000204961.4
|
EFNB1
|
ephrin-B1 |
| chr2_+_65216462 | 0.25 |
ENST00000234256.3
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr5_-_131563501 | 0.24 |
ENST00000401867.1
ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr16_-_84651673 | 0.24 |
ENST00000262428.4
|
COTL1
|
coactosin-like 1 (Dictyostelium) |
| chr11_+_44587141 | 0.24 |
ENST00000227155.4
ENST00000342935.3 ENST00000532544.1 |
CD82
|
CD82 molecule |
| chr12_+_4382917 | 0.24 |
ENST00000261254.3
|
CCND2
|
cyclin D2 |
| chr9_+_71394945 | 0.24 |
ENST00000394264.3
|
FAM122A
|
family with sequence similarity 122A |
| chr8_-_103251274 | 0.24 |
ENST00000251810.3
|
RRM2B
|
ribonucleotide reductase M2 B (TP53 inducible) |
| chr4_-_41216619 | 0.24 |
ENST00000508676.1
ENST00000506352.1 ENST00000295974.8 |
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr6_+_138483058 | 0.24 |
ENST00000251691.4
|
KIAA1244
|
KIAA1244 |
| chr15_+_57884117 | 0.24 |
ENST00000267853.5
|
MYZAP
|
myocardial zonula adherens protein |
| chr2_+_47168313 | 0.23 |
ENST00000319190.5
ENST00000394850.2 ENST00000536057.1 |
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr10_-_74856608 | 0.23 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chr2_-_37899323 | 0.23 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr4_-_89619386 | 0.23 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr4_+_47033345 | 0.23 |
ENST00000295454.3
|
GABRB1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1 |
| chr12_-_42538657 | 0.23 |
ENST00000398675.3
|
GXYLT1
|
glucoside xylosyltransferase 1 |
| chr3_-_125313934 | 0.23 |
ENST00000296220.5
|
OSBPL11
|
oxysterol binding protein-like 11 |
| chr15_-_51058005 | 0.22 |
ENST00000261854.5
|
SPPL2A
|
signal peptide peptidase like 2A |
| chr3_-_178790057 | 0.22 |
ENST00000311417.2
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chr1_-_200992827 | 0.22 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr2_+_26568965 | 0.22 |
ENST00000260585.7
ENST00000447170.1 |
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr9_-_91793675 | 0.22 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr1_-_19283163 | 0.21 |
ENST00000455833.2
|
IFFO2
|
intermediate filament family orphan 2 |
| chr19_+_13135386 | 0.21 |
ENST00000360105.4
ENST00000588228.1 ENST00000591028.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr20_+_45523227 | 0.21 |
ENST00000327619.5
ENST00000357410.3 |
EYA2
|
eyes absent homolog 2 (Drosophila) |
| chr20_-_10654639 | 0.21 |
ENST00000254958.5
|
JAG1
|
jagged 1 |
| chr17_-_49198216 | 0.21 |
ENST00000262013.7
ENST00000357122.4 |
SPAG9
|
sperm associated antigen 9 |
| chr18_-_31803435 | 0.21 |
ENST00000589544.1
ENST00000269185.4 ENST00000261592.5 |
NOL4
|
nucleolar protein 4 |
| chr5_-_27038683 | 0.21 |
ENST00000511822.1
ENST00000231021.4 |
CDH9
|
cadherin 9, type 2 (T1-cadherin) |
| chr11_-_86666427 | 0.21 |
ENST00000531380.1
|
FZD4
|
frizzled family receptor 4 |
| chr5_-_124080203 | 0.21 |
ENST00000504926.1
|
ZNF608
|
zinc finger protein 608 |
| chr20_+_8112824 | 0.21 |
ENST00000378641.3
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific) |
| chr1_+_101361626 | 0.21 |
ENST00000370112.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr1_+_206680879 | 0.21 |
ENST00000355294.4
ENST00000367117.3 |
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr14_+_59104741 | 0.21 |
ENST00000395153.3
ENST00000335867.4 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr2_+_73441350 | 0.21 |
ENST00000389501.4
|
SMYD5
|
SMYD family member 5 |
| chr20_-_58515344 | 0.21 |
ENST00000370996.3
|
PPP1R3D
|
protein phosphatase 1, regulatory subunit 3D |
| chr14_-_74253948 | 0.21 |
ENST00000394071.2
|
ELMSAN1
|
ELM2 and Myb/SANT-like domain containing 1 |
| chr1_+_955448 | 0.20 |
ENST00000379370.2
|
AGRN
|
agrin |
| chr19_-_1863567 | 0.20 |
ENST00000250916.4
|
KLF16
|
Kruppel-like factor 16 |
| chr5_-_95297678 | 0.20 |
ENST00000237853.4
|
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr4_-_100867864 | 0.20 |
ENST00000442697.2
|
DNAJB14
|
DnaJ (Hsp40) homolog, subfamily B, member 14 |
| chr3_-_142166904 | 0.20 |
ENST00000264951.4
|
XRN1
|
5'-3' exoribonuclease 1 |
| chrX_-_33146477 | 0.20 |
ENST00000378677.2
|
DMD
|
dystrophin |
| chr1_+_15943995 | 0.19 |
ENST00000480945.1
|
DDI2
|
DNA-damage inducible 1 homolog 2 (S. cerevisiae) |
| chr21_+_45875354 | 0.19 |
ENST00000291592.4
|
LRRC3
|
leucine rich repeat containing 3 |
| chr4_-_100485143 | 0.19 |
ENST00000394877.3
|
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr10_-_38265517 | 0.19 |
ENST00000302609.7
|
ZNF25
|
zinc finger protein 25 |
| chr1_-_16482554 | 0.19 |
ENST00000358432.5
|
EPHA2
|
EPH receptor A2 |
| chr5_-_43483929 | 0.19 |
ENST00000500337.2
ENST00000506860.1 ENST00000510130.1 ENST00000397080.3 ENST00000512085.1 |
C5orf28
|
chromosome 5 open reading frame 28 |
| chr22_-_50946113 | 0.19 |
ENST00000216080.5
ENST00000474879.2 ENST00000380796.3 |
LMF2
|
lipase maturation factor 2 |
| chr7_-_100076873 | 0.19 |
ENST00000300181.2
|
TSC22D4
|
TSC22 domain family, member 4 |
| chr6_-_43543702 | 0.19 |
ENST00000265351.7
|
XPO5
|
exportin 5 |
| chr11_-_72853091 | 0.19 |
ENST00000311172.7
ENST00000409314.1 |
FCHSD2
|
FCH and double SH3 domains 2 |
| chr3_+_43732362 | 0.18 |
ENST00000458276.2
|
ABHD5
|
abhydrolase domain containing 5 |
| chr3_+_36421826 | 0.18 |
ENST00000273183.3
|
STAC
|
SH3 and cysteine rich domain |
| chr1_+_167691191 | 0.18 |
ENST00000392121.3
ENST00000474859.1 |
MPZL1
|
myelin protein zero-like 1 |
| chr8_-_70747205 | 0.18 |
ENST00000260126.4
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr16_-_80838195 | 0.18 |
ENST00000570137.2
|
CDYL2
|
chromodomain protein, Y-like 2 |
| chr11_+_125462690 | 0.17 |
ENST00000392708.4
ENST00000529196.1 ENST00000531491.1 |
STT3A
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr15_-_23086394 | 0.17 |
ENST00000337435.4
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1 |
| chr6_-_74363803 | 0.17 |
ENST00000355773.5
|
SLC17A5
|
solute carrier family 17 (acidic sugar transporter), member 5 |
| chr6_-_99963252 | 0.17 |
ENST00000392738.2
ENST00000327681.6 ENST00000472914.2 |
USP45
|
ubiquitin specific peptidase 45 |
| chr5_-_131826457 | 0.17 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chr14_+_35515598 | 0.17 |
ENST00000280987.4
|
FAM177A1
|
family with sequence similarity 177, member A1 |
| chr12_+_124196865 | 0.17 |
ENST00000330342.3
|
ATP6V0A2
|
ATPase, H+ transporting, lysosomal V0 subunit a2 |
| chr1_+_155108294 | 0.17 |
ENST00000303343.8
ENST00000368404.4 ENST00000368401.5 |
SLC50A1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr18_+_59854480 | 0.17 |
ENST00000256858.6
ENST00000398130.2 |
KIAA1468
|
KIAA1468 |
| chr10_+_127408263 | 0.17 |
ENST00000337623.3
|
C10orf137
|
erythroid differentiation regulatory factor 1 |
| chr20_+_361261 | 0.17 |
ENST00000217233.3
|
TRIB3
|
tribbles pseudokinase 3 |
| chr10_-_35930219 | 0.17 |
ENST00000374694.1
|
FZD8
|
frizzled family receptor 8 |
| chr20_+_23342783 | 0.17 |
ENST00000544236.1
ENST00000338121.5 ENST00000542987.1 ENST00000424216.1 |
GZF1
|
GDNF-inducible zinc finger protein 1 |
| chr15_-_93199069 | 0.17 |
ENST00000327355.5
|
FAM174B
|
family with sequence similarity 174, member B |
| chr5_-_16936340 | 0.17 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chr2_-_73340146 | 0.17 |
ENST00000258098.6
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I) |
| chr1_+_32645269 | 0.17 |
ENST00000373610.3
|
TXLNA
|
taxilin alpha |
| chr9_+_118916082 | 0.17 |
ENST00000328252.3
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr3_+_119187785 | 0.17 |
ENST00000295588.4
ENST00000476573.1 |
POGLUT1
|
protein O-glucosyltransferase 1 |
| chr9_+_131549483 | 0.16 |
ENST00000372648.5
ENST00000539497.1 |
TBC1D13
|
TBC1 domain family, member 13 |
| chr4_+_95129061 | 0.16 |
ENST00000354268.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr3_+_105085734 | 0.16 |
ENST00000306107.5
|
ALCAM
|
activated leukocyte cell adhesion molecule |
| chr7_+_91875508 | 0.16 |
ENST00000265742.3
|
ANKIB1
|
ankyrin repeat and IBR domain containing 1 |
| chr1_+_205473720 | 0.16 |
ENST00000429964.2
ENST00000506784.1 ENST00000360066.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr4_+_25235597 | 0.16 |
ENST00000264864.6
|
PI4K2B
|
phosphatidylinositol 4-kinase type 2 beta |
| chr4_-_185747188 | 0.16 |
ENST00000507295.1
ENST00000504900.1 ENST00000281455.2 ENST00000454703.2 |
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr2_+_11886710 | 0.16 |
ENST00000256720.2
ENST00000441684.1 ENST00000423495.1 |
LPIN1
|
lipin 1 |
| chr18_+_32073253 | 0.16 |
ENST00000283365.9
ENST00000596745.1 ENST00000315456.6 |
DTNA
|
dystrobrevin, alpha |
| chr5_+_49961727 | 0.16 |
ENST00000505697.2
ENST00000503750.2 ENST00000514342.2 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr2_-_169104651 | 0.16 |
ENST00000355999.4
|
STK39
|
serine threonine kinase 39 |
| chr10_+_71211212 | 0.16 |
ENST00000373290.2
|
TSPAN15
|
tetraspanin 15 |
| chr5_+_109025067 | 0.16 |
ENST00000261483.4
|
MAN2A1
|
mannosidase, alpha, class 2A, member 1 |
| chr12_-_76953284 | 0.15 |
ENST00000547544.1
ENST00000393249.2 |
OSBPL8
|
oxysterol binding protein-like 8 |
| chr9_-_37576226 | 0.15 |
ENST00000432825.2
|
FBXO10
|
F-box protein 10 |
| chr17_-_74449252 | 0.15 |
ENST00000319380.7
|
UBE2O
|
ubiquitin-conjugating enzyme E2O |
| chr5_-_32313019 | 0.15 |
ENST00000280285.5
ENST00000264934.5 |
MTMR12
|
myotubularin related protein 12 |
| chr2_+_26256938 | 0.15 |
ENST00000264710.4
|
RAB10
|
RAB10, member RAS oncogene family |
| chr17_-_1083078 | 0.15 |
ENST00000574266.1
ENST00000302538.5 |
ABR
|
active BCR-related |
| chr10_+_89419370 | 0.15 |
ENST00000361175.4
ENST00000456849.1 |
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr19_-_10444188 | 0.15 |
ENST00000293677.6
|
RAVER1
|
ribonucleoprotein, PTB-binding 1 |
| chr13_-_21476900 | 0.15 |
ENST00000400602.2
ENST00000255305.6 |
XPO4
|
exportin 4 |
| chr18_-_57027194 | 0.15 |
ENST00000251047.5
|
LMAN1
|
lectin, mannose-binding, 1 |
| chr12_-_133338379 | 0.15 |
ENST00000539605.1
|
ANKLE2
|
ankyrin repeat and LEM domain containing 2 |
| chr1_-_222885770 | 0.15 |
ENST00000355727.2
ENST00000340020.6 |
AIDA
|
axin interactor, dorsalization associated |
| chr15_+_90544532 | 0.14 |
ENST00000268154.4
|
ZNF710
|
zinc finger protein 710 |
| chr18_+_19749386 | 0.14 |
ENST00000269216.3
|
GATA6
|
GATA binding protein 6 |
| chr15_+_41952591 | 0.14 |
ENST00000566718.1
ENST00000219905.7 ENST00000389936.4 ENST00000545763.1 |
MGA
|
MGA, MAX dimerization protein |
| chr1_-_101360331 | 0.14 |
ENST00000416479.1
ENST00000370113.3 |
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr7_+_3340989 | 0.14 |
ENST00000404826.2
ENST00000389531.3 |
SDK1
|
sidekick cell adhesion molecule 1 |
| chr4_+_7045042 | 0.14 |
ENST00000310074.7
ENST00000512388.1 |
TADA2B
|
transcriptional adaptor 2B |
| chr10_+_72575643 | 0.14 |
ENST00000373202.3
|
SGPL1
|
sphingosine-1-phosphate lyase 1 |
| chrX_+_103411189 | 0.14 |
ENST00000493442.1
|
FAM199X
|
family with sequence similarity 199, X-linked |
| chr11_-_108093329 | 0.14 |
ENST00000278612.8
|
NPAT
|
nuclear protein, ataxia-telangiectasia locus |
| chr8_-_15095832 | 0.14 |
ENST00000382080.1
|
SGCZ
|
sarcoglycan, zeta |
| chr12_+_113659234 | 0.14 |
ENST00000551096.1
ENST00000551099.1 ENST00000335509.6 ENST00000552897.1 ENST00000550785.1 ENST00000549279.1 |
TPCN1
|
two pore segment channel 1 |
| chr4_-_170192185 | 0.14 |
ENST00000284637.9
|
SH3RF1
|
SH3 domain containing ring finger 1 |
| chr9_-_125675576 | 0.14 |
ENST00000373659.3
|
ZBTB6
|
zinc finger and BTB domain containing 6 |
| chr13_-_28194541 | 0.14 |
ENST00000316334.3
|
LNX2
|
ligand of numb-protein X 2 |
| chr10_-_75173785 | 0.14 |
ENST00000535178.1
ENST00000372921.5 ENST00000372919.4 |
ANXA7
|
annexin A7 |
| chrX_-_128788914 | 0.14 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr12_-_99548270 | 0.14 |
ENST00000546568.1
ENST00000332712.7 ENST00000546960.1 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr5_+_61602055 | 0.14 |
ENST00000381103.2
|
KIF2A
|
kinesin heavy chain member 2A |
| chr11_-_118661828 | 0.14 |
ENST00000264018.4
|
DDX6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr17_-_62658186 | 0.14 |
ENST00000262435.9
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr17_-_1395954 | 0.14 |
ENST00000359786.5
|
MYO1C
|
myosin IC |
| chr10_+_74451883 | 0.13 |
ENST00000373053.3
ENST00000357157.6 |
MCU
|
mitochondrial calcium uniporter |
| chrX_-_109561294 | 0.13 |
ENST00000372059.2
ENST00000262844.5 |
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr15_-_79237433 | 0.13 |
ENST00000220166.5
|
CTSH
|
cathepsin H |
| chr2_+_46769798 | 0.13 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr9_+_97136833 | 0.13 |
ENST00000375344.3
|
HIATL1
|
hippocampus abundant transcript-like 1 |
| chr5_+_151151471 | 0.13 |
ENST00000394123.3
ENST00000543466.1 |
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr9_-_94712434 | 0.13 |
ENST00000375708.3
|
ROR2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr10_-_27443155 | 0.13 |
ENST00000427324.1
ENST00000326799.3 |
YME1L1
|
YME1-like 1 ATPase |
| chrX_+_153607557 | 0.13 |
ENST00000369842.4
ENST00000369835.3 |
EMD
|
emerin |
| chr1_-_72748417 | 0.13 |
ENST00000357731.5
|
NEGR1
|
neuronal growth regulator 1 |
| chr11_-_75062730 | 0.13 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr19_-_3761673 | 0.13 |
ENST00000316757.3
|
APBA3
|
amyloid beta (A4) precursor protein-binding, family A, member 3 |
| chr1_+_28052456 | 0.13 |
ENST00000373954.6
ENST00000419687.2 |
FAM76A
|
family with sequence similarity 76, member A |
| chr13_+_88324870 | 0.13 |
ENST00000325089.6
|
SLITRK5
|
SLIT and NTRK-like family, member 5 |
| chr1_+_151254738 | 0.13 |
ENST00000336715.6
ENST00000324048.5 ENST00000368879.2 |
ZNF687
|
zinc finger protein 687 |
| chr14_-_77279153 | 0.13 |
ENST00000251089.2
|
ANGEL1
|
angel homolog 1 (Drosophila) |
| chr7_-_124405681 | 0.13 |
ENST00000303921.2
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like) |
| chr14_-_104029013 | 0.13 |
ENST00000299204.4
ENST00000557666.1 |
BAG5
|
BCL2-associated athanogene 5 |
| chr20_-_45035223 | 0.13 |
ENST00000450812.1
ENST00000290246.6 ENST00000439931.2 ENST00000396391.1 |
ELMO2
|
engulfment and cell motility 2 |
| chr17_+_72772621 | 0.13 |
ENST00000335464.5
ENST00000417024.2 ENST00000578764.1 ENST00000582773.1 ENST00000582330.1 |
TMEM104
|
transmembrane protein 104 |
| chr16_+_27561449 | 0.13 |
ENST00000261588.4
|
KIAA0556
|
KIAA0556 |
| chr12_+_52404270 | 0.13 |
ENST00000552049.1
ENST00000546756.1 |
GRASP
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein |
| chr4_+_160188889 | 0.13 |
ENST00000264431.4
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr12_-_89918522 | 0.12 |
ENST00000529983.2
|
GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) |
| chrX_+_131157290 | 0.12 |
ENST00000394334.2
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chrX_-_119077695 | 0.12 |
ENST00000371410.3
|
NKAP
|
NFKB activating protein |
| chr13_-_30169807 | 0.12 |
ENST00000380752.5
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr2_+_70142189 | 0.12 |
ENST00000264444.2
|
MXD1
|
MAX dimerization protein 1 |
| chr3_-_27498235 | 0.12 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr20_-_56284816 | 0.12 |
ENST00000395819.3
ENST00000341744.3 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr12_-_93323013 | 0.12 |
ENST00000322349.8
|
EEA1
|
early endosome antigen 1 |
| chr10_-_75910789 | 0.12 |
ENST00000355264.4
|
AP3M1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr12_+_119616447 | 0.12 |
ENST00000281938.2
|
HSPB8
|
heat shock 22kDa protein 8 |
| chr3_+_9404526 | 0.12 |
ENST00000452837.2
ENST00000417036.1 ENST00000419437.1 ENST00000345094.3 ENST00000515662.2 |
THUMPD3
|
THUMP domain containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AAGGCAC
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.6 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 0.3 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.1 | 0.3 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 0.4 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 0.3 | GO:1903410 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.1 | 0.3 | GO:0043132 | NAD transport(GO:0043132) regulation of bleb assembly(GO:1904170) positive regulation of bleb assembly(GO:1904172) |
| 0.1 | 0.3 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.1 | 0.3 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.4 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.2 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.3 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 0.2 | GO:0090427 | regulation of fertilization(GO:0080154) activation of meiosis(GO:0090427) |
| 0.1 | 0.3 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 0.5 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.2 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.1 | 0.2 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.5 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.3 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.2 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.1 | 0.1 | GO:0001743 | optic placode formation(GO:0001743) optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 0.3 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.4 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.2 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.1 | 0.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.2 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.0 | 0.3 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.4 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.4 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.0 | 0.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.4 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.2 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 | 0.1 | GO:0071321 | negative regulation of melanin biosynthetic process(GO:0048022) cellular response to cGMP(GO:0071321) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.2 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.3 | GO:0043634 | polyadenylation-dependent ncRNA catabolic process(GO:0043634) |
| 0.0 | 0.1 | GO:0071109 | superior temporal gyrus development(GO:0071109) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.4 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.1 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.2 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.2 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0032898 | nerve growth factor processing(GO:0032455) neurotrophin production(GO:0032898) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.1 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 0.1 | GO:0042853 | L-alanine metabolic process(GO:0042851) L-alanine catabolic process(GO:0042853) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:1900453 | negative regulation of long term synaptic depression(GO:1900453) regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 0.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.1 | GO:0072560 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.0 | 0.1 | GO:1900756 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.0 | 0.1 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.0 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.5 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.2 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0009644 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.0 | 0.3 | GO:0010891 | regulation of sequestering of triglyceride(GO:0010889) negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:1903722 | negative regulation of exosomal secretion(GO:1903542) regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.2 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.0 | 0.1 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.1 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.0 | 0.1 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.2 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.2 | GO:0045964 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.0 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.0 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.1 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.0 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.0 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.0 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.2 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.2 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.1 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.1 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0001553 | luteinization(GO:0001553) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.1 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.1 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.0 | 0.1 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.7 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 0.4 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.1 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.0 | 0.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.1 | GO:0034681 | integrin alpha11-beta1 complex(GO:0034681) |
| 0.0 | 0.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.0 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.0 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0098560 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.5 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.3 | GO:0005287 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.1 | 0.5 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.2 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.1 | 0.3 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.3 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 0.2 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.3 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.2 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.4 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.1 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.0 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0008478 | pyridoxal kinase activity(GO:0008478) lithium ion binding(GO:0031403) |
| 0.0 | 0.4 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.5 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.3 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0052894 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.0 | 0.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.0 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.7 | PID INSULIN PATHWAY | Insulin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |