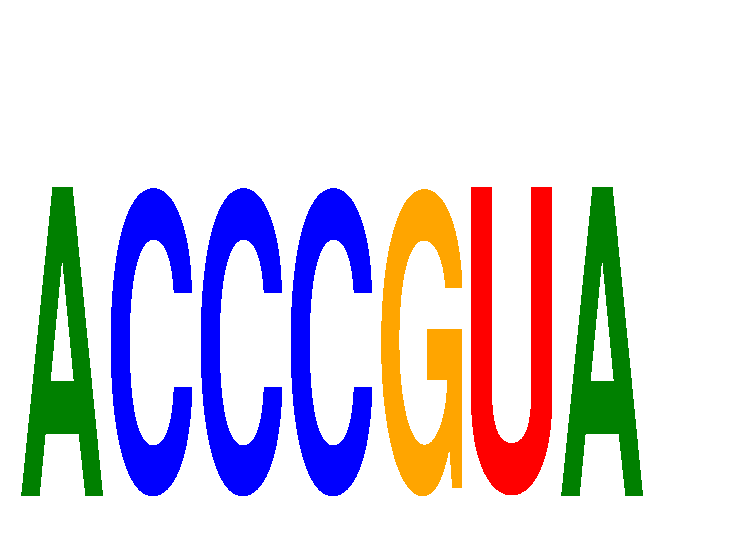Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for ACCCGUA
Z-value: 0.06
miRNA associated with seed ACCCGUA
| Name | miRBASE accession |
|---|---|
|
hsa-miR-99a-5p
|
MIMAT0000097 |
|
hsa-miR-99b-5p
|
MIMAT0000689 |
|
hsa-miR-100-5p
|
MIMAT0000098 |
Activity profile of ACCCGUA motif
Sorted Z-values of ACCCGUA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_12857015 | 0.09 |
ENST00000155926.4
|
TRIB2
|
tribbles pseudokinase 2 |
| chr1_+_65210772 | 0.06 |
ENST00000371072.4
ENST00000294428.3 |
RAVER2
|
ribonucleoprotein, PTB-binding 2 |
| chr4_-_102268628 | 0.05 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr11_+_86748863 | 0.02 |
ENST00000340353.7
|
TMEM135
|
transmembrane protein 135 |
| chr4_+_144434584 | 0.02 |
ENST00000283131.3
|
SMARCA5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr2_-_208634287 | 0.02 |
ENST00000295417.3
|
FZD5
|
frizzled family receptor 5 |
| chr3_+_152017181 | 0.01 |
ENST00000498502.1
ENST00000324196.5 ENST00000545754.1 ENST00000357472.3 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr2_+_33661382 | 0.01 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr22_+_30279144 | 0.01 |
ENST00000401950.2
ENST00000333027.3 ENST00000445401.1 ENST00000323630.5 ENST00000351488.3 |
MTMR3
|
myotubularin related protein 3 |
| chr6_-_75994536 | 0.01 |
ENST00000475111.2
ENST00000230461.6 |
TMEM30A
|
transmembrane protein 30A |
| chr2_+_86947296 | 0.01 |
ENST00000283632.4
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| chr7_+_30323923 | 0.00 |
ENST00000323037.4
|
ZNRF2
|
zinc and ring finger 2 |
Network of associatons between targets according to the STRING database.
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.0 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |