Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for AIRE
Z-value: 0.52
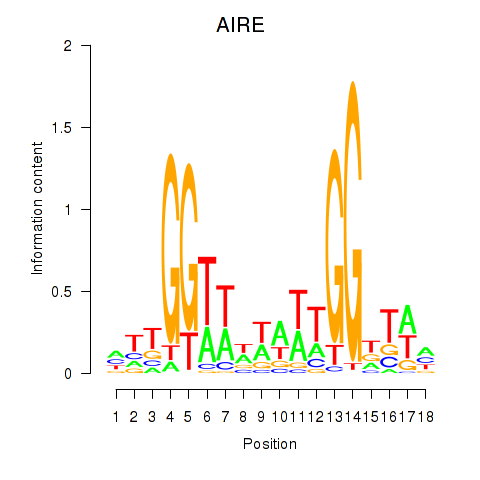
Transcription factors associated with AIRE
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
AIRE
|
ENSG00000160224.12 | autoimmune regulator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| AIRE | hg19_v2_chr21_+_45705752_45705767 | -0.40 | 4.9e-02 | Click! |
Activity profile of AIRE motif
Sorted Z-values of AIRE motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_225811747 | 1.52 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr1_+_79115503 | 0.93 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr8_+_54764346 | 0.85 |
ENST00000297313.3
ENST00000344277.6 |
RGS20
|
regulator of G-protein signaling 20 |
| chr12_+_75874984 | 0.85 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr6_+_127898312 | 0.80 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr1_+_165864821 | 0.79 |
ENST00000470820.1
|
UCK2
|
uridine-cytidine kinase 2 |
| chr12_+_75874580 | 0.68 |
ENST00000456650.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr11_+_35211511 | 0.67 |
ENST00000524922.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr8_+_104384616 | 0.60 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr6_-_32806506 | 0.60 |
ENST00000374897.2
ENST00000452392.2 |
TAP2
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr17_+_77030267 | 0.58 |
ENST00000581774.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr19_-_10491234 | 0.56 |
ENST00000524462.1
ENST00000531836.1 ENST00000525621.1 |
TYK2
|
tyrosine kinase 2 |
| chr5_+_118690466 | 0.55 |
ENST00000503646.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr14_-_107283278 | 0.53 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr1_-_169703203 | 0.49 |
ENST00000333360.7
ENST00000367781.4 ENST00000367782.4 ENST00000367780.4 ENST00000367779.4 |
SELE
|
selectin E |
| chr1_-_154600421 | 0.47 |
ENST00000368471.3
ENST00000292205.5 |
ADAR
|
adenosine deaminase, RNA-specific |
| chr22_+_36649056 | 0.44 |
ENST00000397278.3
ENST00000422706.1 ENST00000426053.1 ENST00000319136.4 |
APOL1
|
apolipoprotein L, 1 |
| chr1_-_243326612 | 0.42 |
ENST00000492145.1
ENST00000490813.1 ENST00000464936.1 |
CEP170
|
centrosomal protein 170kDa |
| chr10_-_106240032 | 0.40 |
ENST00000447860.1
|
RP11-127O4.3
|
RP11-127O4.3 |
| chrM_+_12331 | 0.39 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr4_-_104119528 | 0.35 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr12_+_52643077 | 0.34 |
ENST00000553310.2
ENST00000544024.1 |
KRT86
|
keratin 86 |
| chr17_-_39150385 | 0.34 |
ENST00000391586.1
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr7_-_36764004 | 0.31 |
ENST00000431169.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr9_-_25678856 | 0.31 |
ENST00000358022.3
|
TUSC1
|
tumor suppressor candidate 1 |
| chr7_-_36764142 | 0.31 |
ENST00000258749.5
ENST00000535891.1 |
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr17_-_39334460 | 0.31 |
ENST00000377726.2
|
KRTAP4-2
|
keratin associated protein 4-2 |
| chr16_+_56672571 | 0.30 |
ENST00000290705.8
|
MT1A
|
metallothionein 1A |
| chr21_-_44495919 | 0.30 |
ENST00000398158.1
|
CBS
|
cystathionine-beta-synthase |
| chr1_-_161208013 | 0.29 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr10_+_127661942 | 0.29 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr12_+_10658489 | 0.28 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr10_-_69597810 | 0.27 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr14_-_55658323 | 0.27 |
ENST00000554067.1
ENST00000247191.2 |
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr6_+_26538566 | 0.25 |
ENST00000377575.2
|
HMGN4
|
high mobility group nucleosomal binding domain 4 |
| chr8_+_124780672 | 0.24 |
ENST00000521166.1
ENST00000334705.7 |
FAM91A1
|
family with sequence similarity 91, member A1 |
| chr11_-_5173599 | 0.24 |
ENST00000328942.1
|
OR52A1
|
olfactory receptor, family 52, subfamily A, member 1 |
| chr5_+_122181128 | 0.23 |
ENST00000261369.4
|
SNX24
|
sorting nexin 24 |
| chr13_+_50570019 | 0.23 |
ENST00000442421.1
|
TRIM13
|
tripartite motif containing 13 |
| chr5_+_122181184 | 0.22 |
ENST00000513881.1
|
SNX24
|
sorting nexin 24 |
| chr17_-_38721711 | 0.22 |
ENST00000578085.1
ENST00000246657.2 |
CCR7
|
chemokine (C-C motif) receptor 7 |
| chr14_+_101123580 | 0.21 |
ENST00000556697.1
ENST00000360899.2 ENST00000553623.1 |
LINC00523
|
long intergenic non-protein coding RNA 523 |
| chr12_+_75874460 | 0.21 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr3_-_139199565 | 0.21 |
ENST00000511956.1
|
RBP2
|
retinol binding protein 2, cellular |
| chr3_+_46412345 | 0.21 |
ENST00000292303.4
|
CCR5
|
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
| chr1_+_40840320 | 0.20 |
ENST00000372708.1
|
SMAP2
|
small ArfGAP2 |
| chr17_+_7210294 | 0.20 |
ENST00000336452.7
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr21_-_44495964 | 0.20 |
ENST00000398168.1
ENST00000398165.3 |
CBS
|
cystathionine-beta-synthase |
| chr6_+_22221010 | 0.20 |
ENST00000567753.1
|
RP11-524C21.2
|
RP11-524C21.2 |
| chr14_-_65409438 | 0.20 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr14_-_55658252 | 0.20 |
ENST00000395425.2
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr6_+_160327974 | 0.19 |
ENST00000252660.4
|
MAS1
|
MAS1 oncogene |
| chr17_+_41857793 | 0.19 |
ENST00000449302.3
|
C17orf105
|
chromosome 17 open reading frame 105 |
| chr15_+_85523671 | 0.19 |
ENST00000310298.4
ENST00000557957.1 |
PDE8A
|
phosphodiesterase 8A |
| chrM_+_5824 | 0.19 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr1_-_161207986 | 0.19 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr11_-_8954491 | 0.19 |
ENST00000526227.1
ENST00000525780.1 ENST00000326053.5 |
C11orf16
|
chromosome 11 open reading frame 16 |
| chr19_-_47249679 | 0.18 |
ENST00000263280.6
|
STRN4
|
striatin, calmodulin binding protein 4 |
| chrX_-_101694853 | 0.17 |
ENST00000372749.1
|
NXF2B
|
nuclear RNA export factor 2B |
| chr4_-_57524061 | 0.16 |
ENST00000508121.1
|
HOPX
|
HOP homeobox |
| chr12_-_6579808 | 0.16 |
ENST00000535180.1
ENST00000400911.3 |
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr17_-_42019836 | 0.16 |
ENST00000225992.3
|
PPY
|
pancreatic polypeptide |
| chr12_-_6580094 | 0.15 |
ENST00000361716.3
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr2_-_202508169 | 0.15 |
ENST00000409883.2
|
TMEM237
|
transmembrane protein 237 |
| chrX_-_100307043 | 0.14 |
ENST00000372939.1
ENST00000372935.1 ENST00000372936.3 |
TRMT2B
|
tRNA methyltransferase 2 homolog B (S. cerevisiae) |
| chrX_-_100307076 | 0.14 |
ENST00000338687.7
ENST00000545398.1 ENST00000372931.5 |
TRMT2B
|
tRNA methyltransferase 2 homolog B (S. cerevisiae) |
| chr7_-_149194843 | 0.14 |
ENST00000458143.2
ENST00000340622.3 |
ZNF746
|
zinc finger protein 746 |
| chr4_+_66536248 | 0.13 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr6_-_39290744 | 0.13 |
ENST00000507712.1
|
KCNK16
|
potassium channel, subfamily K, member 16 |
| chr1_-_52456352 | 0.13 |
ENST00000371655.3
|
RAB3B
|
RAB3B, member RAS oncogene family |
| chr11_-_18956556 | 0.12 |
ENST00000302797.3
|
MRGPRX1
|
MAS-related GPR, member X1 |
| chr8_+_105352050 | 0.12 |
ENST00000297581.2
|
DCSTAMP
|
dendrocyte expressed seven transmembrane protein |
| chr16_-_31147020 | 0.12 |
ENST00000568261.1
ENST00000567797.1 ENST00000317508.6 |
PRSS8
|
protease, serine, 8 |
| chr7_+_150498783 | 0.12 |
ENST00000475536.1
ENST00000468689.1 |
TMEM176A
|
transmembrane protein 176A |
| chr2_-_166060571 | 0.12 |
ENST00000360093.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr19_-_35719609 | 0.11 |
ENST00000324675.3
|
FAM187B
|
family with sequence similarity 187, member B |
| chr16_+_50059125 | 0.11 |
ENST00000427478.2
|
CNEP1R1
|
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr9_-_104198042 | 0.11 |
ENST00000374855.4
|
ALDOB
|
aldolase B, fructose-bisphosphate |
| chr5_+_72143988 | 0.11 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr21_+_30502806 | 0.11 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr22_+_25202232 | 0.11 |
ENST00000400358.4
ENST00000400359.4 |
SGSM1
|
small G protein signaling modulator 1 |
| chr11_-_5345582 | 0.11 |
ENST00000328813.2
|
OR51B2
|
olfactory receptor, family 51, subfamily B, member 2 |
| chr12_-_76462713 | 0.11 |
ENST00000552056.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr2_+_207804278 | 0.11 |
ENST00000272852.3
|
CPO
|
carboxypeptidase O |
| chr15_-_81202118 | 0.11 |
ENST00000560560.1
|
RP11-351M8.1
|
Uncharacterized protein |
| chr2_+_201981527 | 0.10 |
ENST00000441224.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr2_-_166060552 | 0.10 |
ENST00000283254.7
ENST00000453007.1 |
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr9_+_100615536 | 0.10 |
ENST00000375123.3
|
FOXE1
|
forkhead box E1 (thyroid transcription factor 2) |
| chr14_-_24911971 | 0.10 |
ENST00000555365.1
ENST00000399395.3 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr9_-_21482312 | 0.10 |
ENST00000448696.3
|
IFNE
|
interferon, epsilon |
| chr20_+_44637526 | 0.10 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr12_+_12202785 | 0.10 |
ENST00000586576.1
ENST00000464885.2 |
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr3_+_193965426 | 0.10 |
ENST00000455821.1
|
RP11-513G11.3
|
RP11-513G11.3 |
| chr12_+_4758264 | 0.09 |
ENST00000266544.5
|
NDUFA9
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa |
| chr12_+_12202774 | 0.09 |
ENST00000589718.1
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr1_+_174670143 | 0.09 |
ENST00000367687.1
ENST00000347255.2 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr6_+_42584847 | 0.09 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr1_+_207669613 | 0.09 |
ENST00000367049.4
ENST00000529814.1 |
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr10_+_82300575 | 0.09 |
ENST00000313455.4
|
SH2D4B
|
SH2 domain containing 4B |
| chr1_+_241815577 | 0.09 |
ENST00000366552.2
ENST00000437684.2 |
WDR64
|
WD repeat domain 64 |
| chr1_+_207669573 | 0.09 |
ENST00000400960.2
ENST00000534202.1 |
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr2_+_179316163 | 0.08 |
ENST00000409117.3
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr2_+_234686976 | 0.08 |
ENST00000389758.3
|
MROH2A
|
maestro heat-like repeat family member 2A |
| chr5_+_32788945 | 0.08 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chr10_-_75682535 | 0.08 |
ENST00000409178.1
|
C10orf55
|
chromosome 10 open reading frame 55 |
| chr10_+_90660832 | 0.08 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr12_-_11139511 | 0.08 |
ENST00000506868.1
|
TAS2R50
|
taste receptor, type 2, member 50 |
| chrX_+_70503433 | 0.07 |
ENST00000276079.8
ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr3_-_33686925 | 0.07 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr1_-_167883327 | 0.07 |
ENST00000476818.2
ENST00000367851.4 ENST00000367848.1 |
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr10_-_101841588 | 0.07 |
ENST00000370418.3
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr14_-_24912047 | 0.07 |
ENST00000553930.1
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr1_-_158300747 | 0.07 |
ENST00000451207.1
|
CD1B
|
CD1b molecule |
| chr7_-_137028498 | 0.07 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr1_-_198990166 | 0.07 |
ENST00000427439.1
|
RP11-16L9.3
|
RP11-16L9.3 |
| chr6_+_42141029 | 0.07 |
ENST00000372958.1
|
GUCA1A
|
guanylate cyclase activator 1A (retina) |
| chr6_+_36839616 | 0.07 |
ENST00000359359.2
ENST00000510325.2 |
C6orf89
|
chromosome 6 open reading frame 89 |
| chr8_+_128748308 | 0.06 |
ENST00000377970.2
|
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr3_+_88199099 | 0.06 |
ENST00000486971.1
|
C3orf38
|
chromosome 3 open reading frame 38 |
| chr12_-_11287243 | 0.06 |
ENST00000539585.1
|
TAS2R30
|
taste receptor, type 2, member 30 |
| chr6_-_31514333 | 0.06 |
ENST00000376151.4
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr11_+_30252549 | 0.06 |
ENST00000254122.3
ENST00000417547.1 |
FSHB
|
follicle stimulating hormone, beta polypeptide |
| chrX_-_49041242 | 0.05 |
ENST00000453382.1
ENST00000540849.1 ENST00000536904.1 ENST00000432913.1 |
PRICKLE3
|
prickle homolog 3 (Drosophila) |
| chr8_-_7220490 | 0.05 |
ENST00000400078.2
|
ZNF705G
|
zinc finger protein 705G |
| chr20_+_21284416 | 0.05 |
ENST00000539513.1
|
XRN2
|
5'-3' exoribonuclease 2 |
| chr16_+_3254247 | 0.05 |
ENST00000304646.2
|
OR1F1
|
olfactory receptor, family 1, subfamily F, member 1 |
| chr3_-_121264848 | 0.05 |
ENST00000264233.5
|
POLQ
|
polymerase (DNA directed), theta |
| chr1_-_145826450 | 0.05 |
ENST00000462900.2
|
GPR89A
|
G protein-coupled receptor 89A |
| chr6_-_31514516 | 0.05 |
ENST00000303892.5
ENST00000483251.1 |
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr4_+_76649797 | 0.05 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr7_-_137028534 | 0.04 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr9_-_4666421 | 0.04 |
ENST00000381895.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr9_+_42717234 | 0.04 |
ENST00000377590.1
|
FOXD4L2
|
forkhead box D4-like 2 |
| chr12_-_11463353 | 0.04 |
ENST00000279575.1
ENST00000535904.1 ENST00000445719.2 |
PRB4
|
proline-rich protein BstNI subfamily 4 |
| chr22_+_42017987 | 0.04 |
ENST00000405506.1
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chrX_-_102941596 | 0.04 |
ENST00000441076.2
ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr11_-_85376121 | 0.03 |
ENST00000527447.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr4_-_120243545 | 0.03 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chrM_+_7586 | 0.03 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr7_+_20686946 | 0.03 |
ENST00000443026.2
ENST00000406935.1 |
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr20_+_54823788 | 0.03 |
ENST00000243911.2
|
MC3R
|
melanocortin 3 receptor |
| chr12_-_59175485 | 0.03 |
ENST00000550678.1
ENST00000552201.1 |
RP11-767I20.1
|
RP11-767I20.1 |
| chr21_+_39644214 | 0.03 |
ENST00000438657.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr1_+_100810575 | 0.03 |
ENST00000542213.1
|
CDC14A
|
cell division cycle 14A |
| chr4_+_78829479 | 0.03 |
ENST00000504901.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr9_-_70178815 | 0.03 |
ENST00000377420.1
|
FOXD4L5
|
forkhead box D4-like 5 |
| chr1_+_206223941 | 0.02 |
ENST00000367126.4
|
AVPR1B
|
arginine vasopressin receptor 1B |
| chr12_+_12223867 | 0.02 |
ENST00000308721.5
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr9_-_70429731 | 0.02 |
ENST00000377413.1
|
FOXD4L4
|
forkhead box D4-like 4 |
| chr6_+_116937636 | 0.02 |
ENST00000368581.4
ENST00000229554.5 ENST00000368580.4 |
RSPH4A
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr15_+_27112948 | 0.02 |
ENST00000555060.1
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr2_+_219840955 | 0.02 |
ENST00000598002.1
ENST00000432733.1 |
LINC00608
|
long intergenic non-protein coding RNA 608 |
| chr3_+_189507460 | 0.02 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr2_-_89568263 | 0.02 |
ENST00000473726.1
|
IGKV1-33
|
immunoglobulin kappa variable 1-33 |
| chr7_-_55606346 | 0.02 |
ENST00000545390.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr2_+_1417228 | 0.02 |
ENST00000382269.3
ENST00000337415.3 ENST00000345913.4 ENST00000346956.3 ENST00000349624.3 ENST00000539820.1 ENST00000329066.4 ENST00000382201.3 |
TPO
|
thyroid peroxidase |
| chr14_-_75330537 | 0.01 |
ENST00000556084.2
ENST00000556489.2 ENST00000445876.1 |
PROX2
|
prospero homeobox 2 |
| chr14_-_106668095 | 0.01 |
ENST00000390606.2
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr15_-_83209210 | 0.01 |
ENST00000561157.1
ENST00000330244.6 |
RPS17L
|
ribosomal protein S17-like |
| chr3_-_133648656 | 0.01 |
ENST00000408895.2
|
C3orf36
|
chromosome 3 open reading frame 36 |
| chr9_-_114361665 | 0.01 |
ENST00000309195.5
|
PTGR1
|
prostaglandin reductase 1 |
| chr22_-_32767017 | 0.01 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr6_+_167525277 | 0.01 |
ENST00000400926.2
|
CCR6
|
chemokine (C-C motif) receptor 6 |
| chr12_-_6579833 | 0.01 |
ENST00000396308.3
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr19_-_16606988 | 0.00 |
ENST00000269881.3
|
CALR3
|
calreticulin 3 |
| chr9_+_117904097 | 0.00 |
ENST00000374016.1
|
DEC1
|
deleted in esophageal cancer 1 |
| chrX_-_153191708 | 0.00 |
ENST00000393721.1
ENST00000370028.3 |
ARHGAP4
|
Rho GTPase activating protein 4 |
| chr2_-_127977654 | 0.00 |
ENST00000409327.1
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr9_-_113761720 | 0.00 |
ENST00000541779.1
ENST00000374430.2 |
LPAR1
|
lysophosphatidic acid receptor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AIRE
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.2 | 0.8 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.2 | 0.5 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.2 | 0.5 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.8 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 1.5 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.6 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 0.7 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.6 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.2 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.1 | 0.2 | GO:0045957 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 0.0 | 0.1 | GO:0034241 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.1 | GO:1904397 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.0 | 0.2 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:0051586 | peptidyl-cysteine methylation(GO:0018125) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.2 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0072186 | metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.5 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.2 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.2 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.6 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 2.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.2 | 0.5 | GO:0070025 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.1 | 0.6 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.1 | 0.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.5 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 0.8 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.2 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.1 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.4 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.2 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.9 | ST G ALPHA I PATHWAY | G alpha i Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |