Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
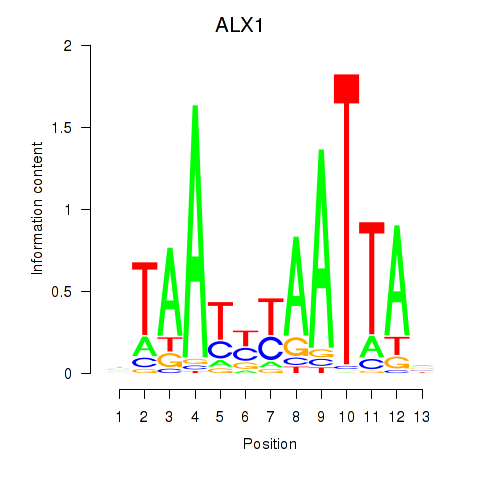
Results for ALX1_ARX
Z-value: 0.33
Transcription factors associated with ALX1_ARX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ALX1
|
ENSG00000180318.3 | ALX homeobox 1 |
|
ARX
|
ENSG00000004848.6 | aristaless related homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ALX1 | hg19_v2_chr12_+_85673868_85673885 | -0.15 | 4.8e-01 | Click! |
| ARX | hg19_v2_chrX_-_25034065_25034088 | -0.00 | 9.8e-01 | Click! |
Activity profile of ALX1_ARX motif
Sorted Z-values of ALX1_ARX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_95236551 | 0.47 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr17_+_39261584 | 0.45 |
ENST00000391415.1
|
KRTAP4-9
|
keratin associated protein 4-9 |
| chr6_-_134639180 | 0.44 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr2_+_162272605 | 0.37 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr6_-_27835357 | 0.35 |
ENST00000331442.3
|
HIST1H1B
|
histone cluster 1, H1b |
| chrX_+_38211777 | 0.32 |
ENST00000039007.4
|
OTC
|
ornithine carbamoyltransferase |
| chr6_+_34204642 | 0.29 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr10_-_31146615 | 0.29 |
ENST00000444692.2
|
ZNF438
|
zinc finger protein 438 |
| chr3_-_164914640 | 0.28 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK-like family, member 3 |
| chr11_+_55029628 | 0.26 |
ENST00000417545.2
|
TRIM48
|
tripartite motif containing 48 |
| chr2_+_152214098 | 0.25 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr18_+_616711 | 0.23 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr2_-_145278475 | 0.22 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr12_-_43833515 | 0.22 |
ENST00000549670.1
ENST00000395541.2 |
ADAMTS20
|
ADAM metallopeptidase with thrombospondin type 1 motif, 20 |
| chr18_+_616672 | 0.21 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr8_+_11961898 | 0.21 |
ENST00000400085.3
|
ZNF705D
|
zinc finger protein 705D |
| chr8_-_10512569 | 0.20 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr11_-_8964580 | 0.20 |
ENST00000325884.1
|
ASCL3
|
achaete-scute family bHLH transcription factor 3 |
| chr12_-_118797475 | 0.20 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr2_+_196313239 | 0.19 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chrX_-_11284095 | 0.16 |
ENST00000303025.6
ENST00000534860.1 |
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr4_-_100356291 | 0.16 |
ENST00000476959.1
ENST00000482593.1 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr3_-_196242233 | 0.16 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr6_+_12007963 | 0.15 |
ENST00000607445.1
|
RP11-456H18.2
|
RP11-456H18.2 |
| chr5_-_160279207 | 0.15 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr3_-_12587055 | 0.15 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr17_+_3118915 | 0.15 |
ENST00000304094.1
|
OR1A1
|
olfactory receptor, family 1, subfamily A, member 1 |
| chr3_+_130569429 | 0.14 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr6_+_12007897 | 0.14 |
ENST00000437559.1
|
RP11-456H18.2
|
RP11-456H18.2 |
| chr3_-_191000172 | 0.14 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr17_-_60885700 | 0.14 |
ENST00000583600.1
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr2_+_102413726 | 0.14 |
ENST00000350878.4
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr17_-_60885659 | 0.14 |
ENST00000311269.5
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr1_+_50459990 | 0.13 |
ENST00000448346.1
|
AL645730.2
|
AL645730.2 |
| chr17_-_39254391 | 0.13 |
ENST00000333822.4
|
KRTAP4-8
|
keratin associated protein 4-8 |
| chr7_+_90339169 | 0.12 |
ENST00000436577.2
|
CDK14
|
cyclin-dependent kinase 14 |
| chr11_-_96076334 | 0.12 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr11_-_40315640 | 0.12 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr10_-_50396425 | 0.12 |
ENST00000374148.1
|
C10orf128
|
chromosome 10 open reading frame 128 |
| chr4_-_177116772 | 0.12 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr12_-_10151773 | 0.12 |
ENST00000298527.6
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1, member B |
| chr1_+_62439037 | 0.12 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr1_-_154600421 | 0.11 |
ENST00000368471.3
ENST00000292205.5 |
ADAR
|
adenosine deaminase, RNA-specific |
| chr1_-_151148442 | 0.11 |
ENST00000441701.1
ENST00000416280.2 |
TMOD4
|
tropomodulin 4 (muscle) |
| chr8_-_61880248 | 0.11 |
ENST00000525556.1
|
AC022182.3
|
AC022182.3 |
| chr12_+_122688090 | 0.10 |
ENST00000324189.4
ENST00000546192.1 |
B3GNT4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chrX_-_19817869 | 0.10 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr4_-_87028478 | 0.10 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr12_+_14572070 | 0.09 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr2_+_119699864 | 0.09 |
ENST00000541757.1
ENST00000412481.1 |
MARCO
|
macrophage receptor with collagenous structure |
| chr17_+_39240459 | 0.09 |
ENST00000391417.4
|
KRTAP4-7
|
keratin associated protein 4-7 |
| chr5_-_59783882 | 0.09 |
ENST00000505507.2
ENST00000502484.2 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr3_-_157824292 | 0.09 |
ENST00000483851.2
|
SHOX2
|
short stature homeobox 2 |
| chr6_+_15401075 | 0.09 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr2_+_119699742 | 0.09 |
ENST00000327097.4
|
MARCO
|
macrophage receptor with collagenous structure |
| chr6_+_3259122 | 0.09 |
ENST00000438998.2
ENST00000380305.4 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr4_-_119759795 | 0.09 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr4_+_69313145 | 0.09 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr18_-_19994830 | 0.09 |
ENST00000525417.1
|
CTAGE1
|
cutaneous T-cell lymphoma-associated antigen 1 |
| chr9_+_124329336 | 0.09 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chrX_+_36254051 | 0.09 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr10_+_69865866 | 0.09 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr1_-_151148492 | 0.08 |
ENST00000295314.4
|
TMOD4
|
tropomodulin 4 (muscle) |
| chr2_+_185463093 | 0.08 |
ENST00000302277.6
|
ZNF804A
|
zinc finger protein 804A |
| chr12_-_118628350 | 0.08 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr2_+_228678550 | 0.08 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr1_-_200379180 | 0.08 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr2_+_12246664 | 0.08 |
ENST00000449986.1
|
AC096559.1
|
AC096559.1 |
| chr5_+_101569696 | 0.08 |
ENST00000597120.1
|
AC008948.1
|
AC008948.1 |
| chr10_+_134150835 | 0.08 |
ENST00000432555.2
|
LRRC27
|
leucine rich repeat containing 27 |
| chr5_+_145826867 | 0.08 |
ENST00000296702.5
ENST00000394421.2 |
TCERG1
|
transcription elongation regulator 1 |
| chr2_+_36923830 | 0.08 |
ENST00000379242.3
ENST00000389975.3 |
VIT
|
vitrin |
| chr17_-_39296739 | 0.08 |
ENST00000345847.4
|
KRTAP4-6
|
keratin associated protein 4-6 |
| chr10_+_18689637 | 0.08 |
ENST00000377315.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr7_+_148287657 | 0.08 |
ENST00000307003.2
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chr4_+_48485341 | 0.07 |
ENST00000273861.4
|
SLC10A4
|
solute carrier family 10, member 4 |
| chr13_-_47471155 | 0.07 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr4_+_88754113 | 0.07 |
ENST00000560249.1
ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr4_-_143227088 | 0.07 |
ENST00000511838.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr8_-_122653630 | 0.07 |
ENST00000303924.4
|
HAS2
|
hyaluronan synthase 2 |
| chr1_+_104615595 | 0.07 |
ENST00000418362.1
|
RP11-364B6.1
|
RP11-364B6.1 |
| chr7_+_90338712 | 0.07 |
ENST00000265741.3
ENST00000406263.1 |
CDK14
|
cyclin-dependent kinase 14 |
| chr1_+_12976450 | 0.07 |
ENST00000361079.2
|
PRAMEF7
|
PRAME family member 7 |
| chr3_-_47934234 | 0.07 |
ENST00000420772.2
|
MAP4
|
microtubule-associated protein 4 |
| chr7_-_111032971 | 0.07 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr17_+_68071458 | 0.07 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr3_-_190167571 | 0.07 |
ENST00000354905.2
|
TMEM207
|
transmembrane protein 207 |
| chr2_+_36923933 | 0.07 |
ENST00000497382.1
ENST00000404084.1 ENST00000379241.3 ENST00000401530.1 |
VIT
|
vitrin |
| chr8_+_77318769 | 0.07 |
ENST00000518732.1
|
RP11-706J10.1
|
long intergenic non-protein coding RNA 1111 |
| chr6_+_45296391 | 0.07 |
ENST00000371436.6
ENST00000576263.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr17_+_68071389 | 0.06 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr15_+_51669444 | 0.06 |
ENST00000396399.2
|
GLDN
|
gliomedin |
| chrX_+_36246735 | 0.06 |
ENST00000378653.3
|
CXorf30
|
chromosome X open reading frame 30 |
| chr5_+_59783941 | 0.06 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr12_-_53601055 | 0.06 |
ENST00000552972.1
ENST00000422257.3 ENST00000267082.5 |
ITGB7
|
integrin, beta 7 |
| chr4_-_143226979 | 0.06 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr6_-_107235287 | 0.06 |
ENST00000436659.1
ENST00000428750.1 ENST00000427903.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr7_+_143701022 | 0.06 |
ENST00000408922.2
|
OR6B1
|
olfactory receptor, family 6, subfamily B, member 1 |
| chr15_+_80733570 | 0.06 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr11_-_7847519 | 0.06 |
ENST00000328375.1
|
OR5P3
|
olfactory receptor, family 5, subfamily P, member 3 |
| chrX_+_149887090 | 0.06 |
ENST00000538506.1
|
MTMR1
|
myotubularin related protein 1 |
| chr6_-_133055815 | 0.06 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr1_-_232651312 | 0.06 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr17_-_3030875 | 0.06 |
ENST00000328890.2
|
OR1G1
|
olfactory receptor, family 1, subfamily G, member 1 |
| chr6_-_32908792 | 0.06 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr8_+_67039131 | 0.05 |
ENST00000315962.4
ENST00000353317.5 |
TRIM55
|
tripartite motif containing 55 |
| chr8_-_57123815 | 0.05 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr14_-_38064198 | 0.05 |
ENST00000250448.2
|
FOXA1
|
forkhead box A1 |
| chr1_+_206557366 | 0.05 |
ENST00000414007.1
ENST00000419187.2 |
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr11_+_62496114 | 0.05 |
ENST00000532583.1
|
TTC9C
|
tetratricopeptide repeat domain 9C |
| chr5_-_87516448 | 0.05 |
ENST00000511218.1
|
TMEM161B
|
transmembrane protein 161B |
| chr11_+_18433840 | 0.05 |
ENST00000541669.1
ENST00000280704.4 |
LDHC
|
lactate dehydrogenase C |
| chr1_-_200379129 | 0.05 |
ENST00000367353.1
|
ZNF281
|
zinc finger protein 281 |
| chr4_+_118955500 | 0.05 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr11_-_111649015 | 0.05 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr17_-_71228357 | 0.05 |
ENST00000583024.1
ENST00000403627.3 ENST00000405159.3 ENST00000581110.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr12_-_53601000 | 0.05 |
ENST00000338737.4
ENST00000549086.2 |
ITGB7
|
integrin, beta 7 |
| chr5_+_40909354 | 0.05 |
ENST00000313164.9
|
C7
|
complement component 7 |
| chr1_-_13390765 | 0.05 |
ENST00000357367.2
|
PRAMEF8
|
PRAME family member 8 |
| chr7_-_25268104 | 0.05 |
ENST00000222674.2
|
NPVF
|
neuropeptide VF precursor |
| chr15_+_42066632 | 0.05 |
ENST00000457542.2
ENST00000221214.6 ENST00000260357.7 ENST00000456763.2 |
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr12_-_125052010 | 0.05 |
ENST00000458234.1
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr6_-_26199499 | 0.04 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr1_+_192127578 | 0.04 |
ENST00000367460.3
|
RGS18
|
regulator of G-protein signaling 18 |
| chr4_-_100356551 | 0.04 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr19_-_38183201 | 0.04 |
ENST00000590008.1
ENST00000358582.4 |
ZNF781
|
zinc finger protein 781 |
| chr3_-_45837959 | 0.04 |
ENST00000353278.4
ENST00000456124.2 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr3_-_194188956 | 0.04 |
ENST00000256031.4
ENST00000446356.1 |
ATP13A3
|
ATPase type 13A3 |
| chr3_-_87325728 | 0.04 |
ENST00000350375.2
|
POU1F1
|
POU class 1 homeobox 1 |
| chr4_+_169013666 | 0.04 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr3_-_45838011 | 0.04 |
ENST00000358525.4
ENST00000413781.1 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr14_-_36988882 | 0.04 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr5_-_27038683 | 0.04 |
ENST00000511822.1
ENST00000231021.4 |
CDH9
|
cadherin 9, type 2 (T1-cadherin) |
| chr3_+_121774202 | 0.04 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr17_-_38911580 | 0.04 |
ENST00000312150.4
|
KRT25
|
keratin 25 |
| chr6_+_26199737 | 0.04 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chr2_+_74685413 | 0.04 |
ENST00000233615.2
|
WBP1
|
WW domain binding protein 1 |
| chr11_-_92931098 | 0.04 |
ENST00000326402.4
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chr1_+_186798073 | 0.04 |
ENST00000367466.3
ENST00000442353.2 |
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr5_-_115872142 | 0.04 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr2_-_55496174 | 0.04 |
ENST00000417363.1
ENST00000412530.1 ENST00000394600.3 ENST00000366137.2 ENST00000420637.1 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr16_+_1728257 | 0.04 |
ENST00000248098.3
ENST00000562684.1 ENST00000561516.1 ENST00000382711.5 ENST00000566742.1 |
HN1L
|
hematological and neurological expressed 1-like |
| chr6_-_66417107 | 0.04 |
ENST00000370621.3
ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS
|
eyes shut homolog (Drosophila) |
| chr18_-_13915530 | 0.03 |
ENST00000327606.3
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr6_-_133055896 | 0.03 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr3_-_101039402 | 0.03 |
ENST00000193391.7
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr18_-_44181442 | 0.03 |
ENST00000398722.4
|
LOXHD1
|
lipoxygenase homology domains 1 |
| chr19_+_7030589 | 0.03 |
ENST00000329753.5
|
MBD3L5
|
methyl-CpG binding domain protein 3-like 5 |
| chr12_-_112123524 | 0.03 |
ENST00000327551.6
|
BRAP
|
BRCA1 associated protein |
| chr4_+_155484155 | 0.03 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr3_-_87325612 | 0.03 |
ENST00000561167.1
ENST00000560656.1 ENST00000344265.3 |
POU1F1
|
POU class 1 homeobox 1 |
| chr4_+_88571429 | 0.03 |
ENST00000339673.6
ENST00000282479.7 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr2_+_58655461 | 0.03 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr5_+_140227357 | 0.03 |
ENST00000378122.3
|
PCDHA9
|
protocadherin alpha 9 |
| chr4_+_74347400 | 0.03 |
ENST00000226355.3
|
AFM
|
afamin |
| chr6_+_31895254 | 0.03 |
ENST00000299367.5
ENST00000442278.2 |
C2
|
complement component 2 |
| chr3_-_3151664 | 0.03 |
ENST00000256452.3
ENST00000311981.8 ENST00000430514.2 ENST00000456302.1 |
IL5RA
|
interleukin 5 receptor, alpha |
| chr3_+_111393501 | 0.03 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr12_+_122241928 | 0.03 |
ENST00000604567.1
ENST00000542440.1 |
SETD1B
|
SET domain containing 1B |
| chr2_+_75873902 | 0.03 |
ENST00000393909.2
ENST00000358788.6 ENST00000409374.1 |
MRPL19
|
mitochondrial ribosomal protein L19 |
| chr4_+_113568207 | 0.03 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr6_-_137314371 | 0.03 |
ENST00000432330.1
ENST00000418699.1 |
RP11-55K22.5
|
RP11-55K22.5 |
| chr3_-_186262166 | 0.03 |
ENST00000307944.5
|
CRYGS
|
crystallin, gamma S |
| chr6_-_26199471 | 0.03 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chrX_+_107288197 | 0.03 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chrX_-_15332665 | 0.03 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr10_+_89420706 | 0.03 |
ENST00000427144.2
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chrX_-_15619076 | 0.02 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr4_-_83765613 | 0.02 |
ENST00000503937.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr3_+_155860751 | 0.02 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chrX_+_107288239 | 0.02 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chrX_+_16668278 | 0.02 |
ENST00000380200.3
|
S100G
|
S100 calcium binding protein G |
| chrX_+_119737806 | 0.02 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr3_-_32544900 | 0.02 |
ENST00000205636.3
|
CMTM6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr10_-_75226166 | 0.02 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr9_+_42671887 | 0.02 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr15_+_44092784 | 0.02 |
ENST00000458412.1
|
HYPK
|
huntingtin interacting protein K |
| chr4_+_54927213 | 0.02 |
ENST00000595906.1
|
AC110792.1
|
HCG2027126; Uncharacterized protein |
| chr9_+_77230499 | 0.02 |
ENST00000396204.2
|
RORB
|
RAR-related orphan receptor B |
| chr2_+_161993412 | 0.02 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr8_-_86253888 | 0.02 |
ENST00000522389.1
ENST00000432364.2 ENST00000517618.1 |
CA1
|
carbonic anhydrase I |
| chr6_-_150067696 | 0.02 |
ENST00000340413.2
ENST00000367403.3 |
NUP43
|
nucleoporin 43kDa |
| chr21_-_43346790 | 0.02 |
ENST00000329623.7
|
C2CD2
|
C2 calcium-dependent domain containing 2 |
| chrX_-_48858667 | 0.02 |
ENST00000376423.4
ENST00000376441.1 |
GRIPAP1
|
GRIP1 associated protein 1 |
| chr10_-_29923893 | 0.02 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr1_+_158323755 | 0.02 |
ENST00000368157.1
ENST00000368156.1 ENST00000368155.3 ENST00000368154.1 ENST00000368160.3 ENST00000368161.3 |
CD1E
|
CD1e molecule |
| chr5_+_137203465 | 0.02 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chrX_-_48858630 | 0.02 |
ENST00000376425.3
ENST00000376444.3 |
GRIPAP1
|
GRIP1 associated protein 1 |
| chr10_-_102989551 | 0.02 |
ENST00000370193.2
|
LBX1
|
ladybird homeobox 1 |
| chr9_-_95298314 | 0.02 |
ENST00000344604.5
ENST00000375540.1 |
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr11_+_113779704 | 0.02 |
ENST00000537778.1
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B, ionotropic |
| chr4_+_156824840 | 0.02 |
ENST00000536354.2
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr8_+_7353368 | 0.02 |
ENST00000355602.2
|
DEFB107B
|
defensin, beta 107B |
| chr11_-_124190184 | 0.02 |
ENST00000357438.2
|
OR8D2
|
olfactory receptor, family 8, subfamily D, member 2 |
| chr9_+_44867571 | 0.02 |
ENST00000377548.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr14_-_81893734 | 0.02 |
ENST00000555447.1
|
STON2
|
stonin 2 |
| chr4_-_89951028 | 0.01 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr4_-_176733897 | 0.01 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr4_+_95376396 | 0.01 |
ENST00000508216.1
ENST00000514743.1 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr19_+_7049332 | 0.01 |
ENST00000381393.3
|
MBD3L2
|
methyl-CpG binding domain protein 3-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ALX1_ARX
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 0.1 | 0.3 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.1 | 0.2 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.4 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.3 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.4 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.3 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.1 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.4 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.1 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.0 | 0.1 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.1 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.0 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.3 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.0 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |