Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
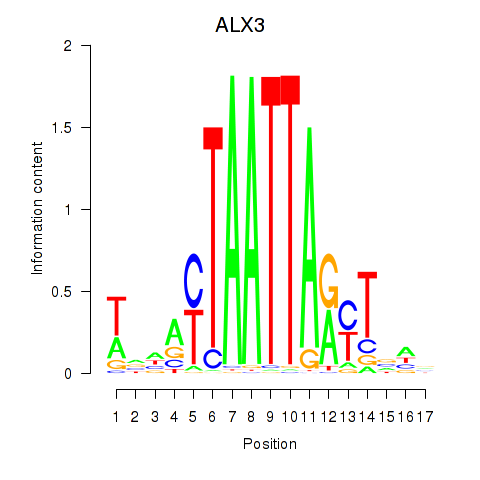
Results for ALX3
Z-value: 0.56
Transcription factors associated with ALX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ALX3
|
ENSG00000156150.6 | ALX homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ALX3 | hg19_v2_chr1_-_110613276_110613322 | -0.15 | 4.9e-01 | Click! |
Activity profile of ALX3 motif
Sorted Z-values of ALX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_86525299 | 2.04 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr10_+_91152303 | 0.90 |
ENST00000371804.3
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr10_+_6779326 | 0.86 |
ENST00000417112.1
|
RP11-554I8.2
|
RP11-554I8.2 |
| chr6_+_127898312 | 0.77 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr2_-_225811747 | 0.74 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr16_-_28621353 | 0.62 |
ENST00000567512.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr20_+_31805131 | 0.60 |
ENST00000375454.3
ENST00000375452.3 |
BPIFA3
|
BPI fold containing family A, member 3 |
| chr15_+_94899183 | 0.55 |
ENST00000557742.1
|
MCTP2
|
multiple C2 domains, transmembrane 2 |
| chr11_+_5710919 | 0.49 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr8_-_116504448 | 0.45 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr6_+_26402465 | 0.45 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr6_+_26402517 | 0.44 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr1_+_62439037 | 0.44 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr20_-_56265680 | 0.39 |
ENST00000414037.1
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr18_+_29171689 | 0.37 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr17_-_2996290 | 0.36 |
ENST00000331459.1
|
OR1D2
|
olfactory receptor, family 1, subfamily D, member 2 |
| chr1_-_67266939 | 0.35 |
ENST00000304526.2
|
INSL5
|
insulin-like 5 |
| chr1_-_150738261 | 0.34 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr2_+_68962014 | 0.33 |
ENST00000467265.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr7_-_111032971 | 0.31 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr2_-_220264703 | 0.31 |
ENST00000519905.1
ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP
|
aspartyl aminopeptidase |
| chrX_-_21676442 | 0.31 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr11_-_5323226 | 0.31 |
ENST00000380224.1
|
OR51B4
|
olfactory receptor, family 51, subfamily B, member 4 |
| chr3_-_191000172 | 0.30 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr1_+_28261533 | 0.29 |
ENST00000411604.1
ENST00000373888.4 |
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr1_+_28261492 | 0.29 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr4_+_95128748 | 0.29 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr4_+_89206076 | 0.28 |
ENST00000500009.2
|
RP11-10L7.1
|
RP11-10L7.1 |
| chr9_-_117111222 | 0.28 |
ENST00000374079.4
|
AKNA
|
AT-hook transcription factor |
| chr19_-_3557570 | 0.28 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr5_-_20575959 | 0.28 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr2_-_73520667 | 0.27 |
ENST00000545030.1
ENST00000436467.2 |
EGR4
|
early growth response 4 |
| chr1_+_28261621 | 0.27 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr2_+_103035102 | 0.26 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr22_+_17956618 | 0.26 |
ENST00000262608.8
|
CECR2
|
cat eye syndrome chromosome region, candidate 2 |
| chr6_+_26440700 | 0.26 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr11_+_35201826 | 0.26 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr5_-_35938674 | 0.26 |
ENST00000397366.1
ENST00000513623.1 ENST00000514524.1 ENST00000397367.2 |
CAPSL
|
calcyphosine-like |
| chr11_+_55594695 | 0.25 |
ENST00000378397.1
|
OR5L2
|
olfactory receptor, family 5, subfamily L, member 2 |
| chr14_-_107049312 | 0.25 |
ENST00000390627.2
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr1_+_144989309 | 0.24 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr12_+_26348246 | 0.24 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr16_-_28634874 | 0.24 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr3_+_40518599 | 0.24 |
ENST00000314686.5
ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619
|
zinc finger protein 619 |
| chr14_+_22977587 | 0.23 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chr17_-_72772462 | 0.23 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr17_+_18086392 | 0.23 |
ENST00000541285.1
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli) |
| chr7_+_138145076 | 0.22 |
ENST00000343526.4
|
TRIM24
|
tripartite motif containing 24 |
| chr3_+_185431080 | 0.22 |
ENST00000296270.1
|
C3orf65
|
chromosome 3 open reading frame 65 |
| chr4_-_159956333 | 0.22 |
ENST00000434826.2
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr4_+_5527117 | 0.22 |
ENST00000505296.1
|
C4orf6
|
chromosome 4 open reading frame 6 |
| chr7_-_92777606 | 0.21 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr19_-_44388116 | 0.21 |
ENST00000587539.1
|
ZNF404
|
zinc finger protein 404 |
| chr1_+_115572415 | 0.20 |
ENST00000256592.1
|
TSHB
|
thyroid stimulating hormone, beta |
| chr1_+_225600404 | 0.20 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr14_+_32798462 | 0.19 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr12_+_26348429 | 0.19 |
ENST00000242729.2
|
SSPN
|
sarcospan |
| chr11_+_7618413 | 0.19 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr7_-_87342564 | 0.19 |
ENST00000265724.3
ENST00000416177.1 |
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr2_-_136678123 | 0.19 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr9_+_105757590 | 0.18 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr17_-_3030875 | 0.18 |
ENST00000328890.2
|
OR1G1
|
olfactory receptor, family 1, subfamily G, member 1 |
| chr17_-_19015945 | 0.18 |
ENST00000573866.2
|
SNORD3D
|
small nucleolar RNA, C/D box 3D |
| chr16_+_31885079 | 0.18 |
ENST00000300870.10
ENST00000394846.3 |
ZNF267
|
zinc finger protein 267 |
| chr20_+_57594309 | 0.18 |
ENST00000217133.1
|
TUBB1
|
tubulin, beta 1 class VI |
| chr11_+_100862811 | 0.18 |
ENST00000303130.2
|
TMEM133
|
transmembrane protein 133 |
| chr2_+_68961934 | 0.17 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr2_+_68961905 | 0.17 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr6_-_87804815 | 0.17 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr16_+_31271274 | 0.17 |
ENST00000287497.8
ENST00000544665.3 |
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr10_+_24755416 | 0.17 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr12_-_23737534 | 0.16 |
ENST00000396007.2
|
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr14_+_74034310 | 0.16 |
ENST00000538782.1
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr16_-_28608424 | 0.16 |
ENST00000335715.4
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr2_+_102953608 | 0.16 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr2_-_43266680 | 0.16 |
ENST00000425212.1
ENST00000422351.1 ENST00000449766.1 |
AC016735.2
|
AC016735.2 |
| chr2_-_228497888 | 0.16 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr2_-_89385283 | 0.16 |
ENST00000390252.2
|
IGKV3-15
|
immunoglobulin kappa variable 3-15 |
| chr17_-_64225508 | 0.16 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr16_-_28608364 | 0.16 |
ENST00000533150.1
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr11_+_71900572 | 0.16 |
ENST00000312293.4
|
FOLR1
|
folate receptor 1 (adult) |
| chr9_+_95709733 | 0.16 |
ENST00000375482.3
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr1_-_202897724 | 0.16 |
ENST00000435533.3
ENST00000367258.1 |
KLHL12
|
kelch-like family member 12 |
| chr6_-_82957433 | 0.16 |
ENST00000306270.7
|
IBTK
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chrX_+_107288239 | 0.15 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr3_+_140396881 | 0.15 |
ENST00000286349.3
|
TRIM42
|
tripartite motif containing 42 |
| chr5_+_176811431 | 0.15 |
ENST00000512593.1
ENST00000324417.5 |
SLC34A1
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 1 |
| chr11_+_71900703 | 0.15 |
ENST00000393681.2
|
FOLR1
|
folate receptor 1 (adult) |
| chr20_-_50418972 | 0.15 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr20_-_50418947 | 0.15 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chrX_+_56100757 | 0.15 |
ENST00000433279.1
|
AL353698.1
|
Uncharacterized protein |
| chr17_+_19091325 | 0.15 |
ENST00000584923.1
|
SNORD3A
|
small nucleolar RNA, C/D box 3A |
| chr4_-_185275104 | 0.15 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr2_+_90273679 | 0.15 |
ENST00000423080.2
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr2_+_145425573 | 0.15 |
ENST00000600064.1
ENST00000597670.1 ENST00000414256.1 ENST00000599187.1 ENST00000451774.1 ENST00000599072.1 ENST00000596589.1 ENST00000597893.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr1_+_47533160 | 0.15 |
ENST00000334194.3
|
CYP4Z1
|
cytochrome P450, family 4, subfamily Z, polypeptide 1 |
| chr16_-_28621298 | 0.14 |
ENST00000566189.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr4_+_110736659 | 0.14 |
ENST00000394631.3
ENST00000226796.6 |
GAR1
|
GAR1 ribonucleoprotein |
| chr12_-_86650045 | 0.14 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr14_-_67878917 | 0.14 |
ENST00000216446.4
|
PLEK2
|
pleckstrin 2 |
| chr7_+_156433573 | 0.14 |
ENST00000311822.8
|
RNF32
|
ring finger protein 32 |
| chr2_+_90077680 | 0.14 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr10_-_50396407 | 0.14 |
ENST00000374153.2
ENST00000374151.3 |
C10orf128
|
chromosome 10 open reading frame 128 |
| chr17_-_39341594 | 0.13 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr10_-_50396425 | 0.13 |
ENST00000374148.1
|
C10orf128
|
chromosome 10 open reading frame 128 |
| chr15_-_98417780 | 0.13 |
ENST00000503874.3
|
LINC00923
|
long intergenic non-protein coding RNA 923 |
| chr20_+_33134619 | 0.13 |
ENST00000374837.3
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha |
| chr10_+_35894338 | 0.13 |
ENST00000321660.1
|
GJD4
|
gap junction protein, delta 4, 40.1kDa |
| chrX_-_77225135 | 0.13 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr3_+_119298523 | 0.13 |
ENST00000357003.3
|
ADPRH
|
ADP-ribosylarginine hydrolase |
| chr4_+_66536248 | 0.13 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr7_-_121944491 | 0.13 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr7_+_117864708 | 0.12 |
ENST00000357099.4
ENST00000265224.4 ENST00000486422.1 ENST00000417525.1 |
ANKRD7
|
ankyrin repeat domain 7 |
| chr3_+_51851612 | 0.12 |
ENST00000456080.1
|
IQCF3
|
IQ motif containing F3 |
| chr2_+_135596180 | 0.12 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr7_+_120628731 | 0.12 |
ENST00000310396.5
|
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr20_-_50722183 | 0.12 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr16_-_28621312 | 0.12 |
ENST00000314752.7
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr11_+_6897856 | 0.12 |
ENST00000379829.2
|
OR10A4
|
olfactory receptor, family 10, subfamily A, member 4 |
| chr6_-_7313381 | 0.12 |
ENST00000489567.1
ENST00000479365.1 ENST00000462112.1 ENST00000397511.2 ENST00000534851.1 ENST00000474597.1 ENST00000244763.4 |
SSR1
|
signal sequence receptor, alpha |
| chr11_+_60163775 | 0.12 |
ENST00000300187.6
ENST00000395005.2 |
MS4A14
|
membrane-spanning 4-domains, subfamily A, member 14 |
| chr1_-_92952433 | 0.12 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr4_+_37455536 | 0.11 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr2_-_198540719 | 0.11 |
ENST00000295049.4
|
RFTN2
|
raftlin family member 2 |
| chr2_+_187371440 | 0.11 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr4_+_88720698 | 0.11 |
ENST00000226284.5
|
IBSP
|
integrin-binding sialoprotein |
| chr2_-_74619152 | 0.11 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chr4_+_5526883 | 0.11 |
ENST00000195455.2
|
C4orf6
|
chromosome 4 open reading frame 6 |
| chr2_+_135596106 | 0.11 |
ENST00000356140.5
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr12_-_122985067 | 0.11 |
ENST00000540586.1
ENST00000543897.1 |
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr19_+_17638041 | 0.10 |
ENST00000601861.1
|
FAM129C
|
family with sequence similarity 129, member C |
| chr15_+_64680003 | 0.10 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr5_+_140762268 | 0.10 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr11_+_2405833 | 0.10 |
ENST00000527343.1
ENST00000464784.2 |
CD81
|
CD81 molecule |
| chr6_+_154360476 | 0.10 |
ENST00000428397.2
|
OPRM1
|
opioid receptor, mu 1 |
| chr14_-_106668095 | 0.10 |
ENST00000390606.2
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chrX_+_84258832 | 0.10 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr21_-_22175341 | 0.10 |
ENST00000416768.1
ENST00000452561.1 ENST00000419299.1 ENST00000437238.1 |
LINC00320
|
long intergenic non-protein coding RNA 320 |
| chr3_-_160823040 | 0.10 |
ENST00000484127.1
ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr15_+_58702742 | 0.09 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr14_+_32798547 | 0.09 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr2_-_74618964 | 0.09 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr15_+_58724184 | 0.09 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr8_+_30244580 | 0.09 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr9_-_115095123 | 0.09 |
ENST00000458258.1
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr6_+_158733692 | 0.09 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr8_-_12612962 | 0.09 |
ENST00000398246.3
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr3_+_121774202 | 0.09 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr16_+_31724552 | 0.08 |
ENST00000539915.1
ENST00000316491.9 ENST00000399681.3 ENST00000398696.3 ENST00000534369.1 |
ZNF720
|
zinc finger protein 720 |
| chr7_-_44580861 | 0.08 |
ENST00000546276.1
ENST00000289547.4 ENST00000381160.3 ENST00000423141.1 |
NPC1L1
|
NPC1-like 1 |
| chr15_-_89755034 | 0.08 |
ENST00000563254.1
|
RLBP1
|
retinaldehyde binding protein 1 |
| chrX_+_108779004 | 0.08 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr13_+_78315295 | 0.08 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr1_-_185597619 | 0.08 |
ENST00000608417.1
ENST00000436955.1 |
GS1-204I12.1
|
GS1-204I12.1 |
| chr3_-_101039402 | 0.08 |
ENST00000193391.7
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr14_+_39736582 | 0.08 |
ENST00000556148.1
ENST00000348007.3 |
CTAGE5
|
CTAGE family, member 5 |
| chr1_-_24126051 | 0.08 |
ENST00000445705.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr2_-_231860596 | 0.08 |
ENST00000441063.1
ENST00000434094.1 ENST00000418330.1 ENST00000457803.1 ENST00000414876.1 ENST00000446741.1 ENST00000426904.1 |
AC105344.2
|
SPATA3 antisense RNA 1 (head to head) |
| chr2_-_3521518 | 0.08 |
ENST00000382093.5
|
ADI1
|
acireductone dioxygenase 1 |
| chr5_-_98262240 | 0.08 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chrY_-_6740649 | 0.08 |
ENST00000383036.1
ENST00000383037.4 |
AMELY
|
amelogenin, Y-linked |
| chr7_-_2883928 | 0.08 |
ENST00000275364.3
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr3_-_160823158 | 0.08 |
ENST00000392779.2
ENST00000392780.1 ENST00000494173.1 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr4_-_103749205 | 0.07 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_+_71859156 | 0.07 |
ENST00000286648.5
ENST00000504730.1 ENST00000504952.1 |
DCK
|
deoxycytidine kinase |
| chr8_+_55528627 | 0.07 |
ENST00000220676.1
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant) |
| chrX_+_107288197 | 0.07 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr10_+_18549645 | 0.07 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr15_+_75970672 | 0.07 |
ENST00000435356.1
|
AC105020.1
|
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chr9_+_470288 | 0.07 |
ENST00000382303.1
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr14_-_106552755 | 0.07 |
ENST00000390600.2
|
IGHV3-9
|
immunoglobulin heavy variable 3-9 |
| chr8_-_93978357 | 0.07 |
ENST00000522925.1
ENST00000522903.1 ENST00000537541.1 ENST00000518748.1 ENST00000519069.1 ENST00000521988.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr12_+_18414446 | 0.07 |
ENST00000433979.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
| chr6_-_33860521 | 0.07 |
ENST00000525746.1
ENST00000531046.1 |
LINC01016
|
long intergenic non-protein coding RNA 1016 |
| chr8_-_30670384 | 0.06 |
ENST00000221138.4
ENST00000518243.1 |
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr6_-_105307711 | 0.06 |
ENST00000519645.1
ENST00000262903.4 ENST00000369125.2 |
HACE1
|
HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 |
| chr11_-_62521614 | 0.06 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr4_-_120243545 | 0.06 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr19_+_42580274 | 0.06 |
ENST00000359044.4
|
ZNF574
|
zinc finger protein 574 |
| chr11_+_92702886 | 0.06 |
ENST00000257068.2
ENST00000528076.1 |
MTNR1B
|
melatonin receptor 1B |
| chr11_-_107729887 | 0.06 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr4_-_70518941 | 0.06 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr19_+_17638059 | 0.06 |
ENST00000599164.1
ENST00000449408.2 ENST00000600871.1 ENST00000599124.1 |
FAM129C
|
family with sequence similarity 129, member C |
| chr8_-_623547 | 0.06 |
ENST00000522893.1
|
ERICH1
|
glutamate-rich 1 |
| chr14_+_55493920 | 0.06 |
ENST00000395472.2
ENST00000555846.1 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr20_-_29978383 | 0.05 |
ENST00000339144.3
ENST00000376321.3 |
DEFB119
|
defensin, beta 119 |
| chr1_-_24306835 | 0.05 |
ENST00000484146.2
|
SRSF10
|
serine/arginine-rich splicing factor 10 |
| chr1_-_24306768 | 0.05 |
ENST00000374453.3
ENST00000453840.3 |
SRSF10
|
serine/arginine-rich splicing factor 10 |
| chr9_-_116065551 | 0.05 |
ENST00000297894.5
|
RNF183
|
ring finger protein 183 |
| chr3_+_148709128 | 0.05 |
ENST00000345003.4
ENST00000296048.6 ENST00000483267.1 |
GYG1
|
glycogenin 1 |
| chr5_+_140201183 | 0.05 |
ENST00000529619.1
ENST00000529859.1 ENST00000378126.3 |
PCDHA5
|
protocadherin alpha 5 |
| chr2_+_145425534 | 0.05 |
ENST00000432608.1
ENST00000597655.1 ENST00000598659.1 ENST00000600679.1 ENST00000601277.1 ENST00000451027.1 ENST00000445791.1 ENST00000596540.1 ENST00000596230.1 ENST00000594471.1 ENST00000598248.1 ENST00000597469.1 ENST00000431734.1 ENST00000595686.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr1_-_207226313 | 0.05 |
ENST00000367084.1
|
YOD1
|
YOD1 deubiquitinase |
| chr14_+_55494323 | 0.05 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr8_+_50824233 | 0.05 |
ENST00000522124.1
|
SNTG1
|
syntrophin, gamma 1 |
| chr1_-_150669604 | 0.05 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr4_-_103749313 | 0.05 |
ENST00000394803.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_-_103749105 | 0.05 |
ENST00000394801.4
ENST00000394804.2 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr12_+_8666126 | 0.04 |
ENST00000299665.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr9_+_124329336 | 0.04 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of ALX3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 0.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.3 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.1 | 0.8 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.2 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 1.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.7 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.2 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.3 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.2 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.3 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.9 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.2 | GO:1903961 | positive regulation of anion channel activity(GO:1901529) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.0 | 0.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.3 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.0 | GO:0061346 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 | 0.2 | GO:0034392 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:1990168 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.2 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.2 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.3 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.3 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.3 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.2 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.3 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.0 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.0 | GO:0060187 | cell pole(GO:0060187) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.1 | 0.3 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.1 | 0.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.2 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 0.2 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 0.2 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 0.2 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.2 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 2.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |