Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
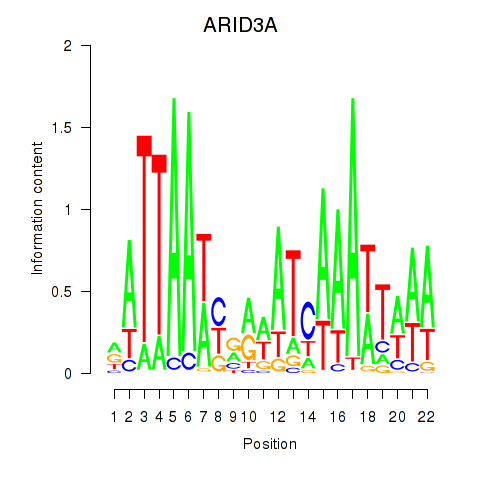
Results for ARID3A
Z-value: 0.64
Transcription factors associated with ARID3A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID3A
|
ENSG00000116017.6 | AT-rich interaction domain 3A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID3A | hg19_v2_chr19_+_926000_926046 | 0.37 | 7.2e-02 | Click! |
Activity profile of ARID3A motif
Sorted Z-values of ARID3A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_76957214 | 4.25 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr12_+_113344582 | 2.44 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr1_+_145438469 | 1.85 |
ENST00000369317.4
|
TXNIP
|
thioredoxin interacting protein |
| chr13_-_33780133 | 1.11 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr7_-_139763521 | 0.97 |
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr12_+_97306295 | 0.84 |
ENST00000457368.2
|
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr4_-_169239921 | 0.83 |
ENST00000514995.1
ENST00000393743.3 |
DDX60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr4_+_70916119 | 0.82 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr8_+_11961898 | 0.81 |
ENST00000400085.3
|
ZNF705D
|
zinc finger protein 705D |
| chr10_+_91092241 | 0.79 |
ENST00000371811.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr6_+_132891461 | 0.75 |
ENST00000275198.1
|
TAAR6
|
trace amine associated receptor 6 |
| chr13_-_102068706 | 0.73 |
ENST00000251127.6
|
NALCN
|
sodium leak channel, non-selective |
| chr4_-_80994210 | 0.70 |
ENST00000403729.2
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr5_-_59783882 | 0.64 |
ENST00000505507.2
ENST00000502484.2 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr10_+_111765562 | 0.62 |
ENST00000360162.3
|
ADD3
|
adducin 3 (gamma) |
| chr3_-_52804872 | 0.62 |
ENST00000535191.1
ENST00000461689.1 ENST00000383721.4 ENST00000233027.5 |
NEK4
|
NIMA-related kinase 4 |
| chr18_-_19994830 | 0.62 |
ENST00000525417.1
|
CTAGE1
|
cutaneous T-cell lymphoma-associated antigen 1 |
| chr12_-_71182695 | 0.56 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr2_+_119699742 | 0.55 |
ENST00000327097.4
|
MARCO
|
macrophage receptor with collagenous structure |
| chr2_+_119699864 | 0.55 |
ENST00000541757.1
ENST00000412481.1 |
MARCO
|
macrophage receptor with collagenous structure |
| chr12_-_68696652 | 0.53 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr11_+_35201826 | 0.53 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr11_-_5207612 | 0.52 |
ENST00000380367.1
|
OR52A1
|
olfactory receptor, family 52, subfamily A, member 1 |
| chr8_-_62559366 | 0.50 |
ENST00000522919.1
|
ASPH
|
aspartate beta-hydroxylase |
| chr11_-_5345582 | 0.50 |
ENST00000328813.2
|
OR51B2
|
olfactory receptor, family 51, subfamily B, member 2 |
| chr12_+_133757995 | 0.49 |
ENST00000536435.2
ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr3_-_169487617 | 0.49 |
ENST00000330368.2
|
ACTRT3
|
actin-related protein T3 |
| chr1_-_154600421 | 0.49 |
ENST00000368471.3
ENST00000292205.5 |
ADAR
|
adenosine deaminase, RNA-specific |
| chr3_+_169629354 | 0.49 |
ENST00000428432.2
ENST00000335556.3 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr3_+_142442841 | 0.48 |
ENST00000476941.1
ENST00000273482.6 |
TRPC1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr6_-_29055090 | 0.48 |
ENST00000377173.2
|
OR2B3
|
olfactory receptor, family 2, subfamily B, member 3 |
| chr4_+_74301880 | 0.47 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr4_-_80994619 | 0.46 |
ENST00000404191.1
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr4_-_80994471 | 0.46 |
ENST00000295465.4
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr1_-_55230165 | 0.45 |
ENST00000371279.3
|
PARS2
|
prolyl-tRNA synthetase 2, mitochondrial (putative) |
| chr4_+_113568207 | 0.42 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr10_-_100995603 | 0.42 |
ENST00000370552.3
ENST00000370549.1 |
HPSE2
|
heparanase 2 |
| chr15_+_64680003 | 0.42 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr2_+_234959376 | 0.42 |
ENST00000425558.1
|
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr9_+_118916082 | 0.41 |
ENST00000328252.3
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr16_+_76587314 | 0.40 |
ENST00000563764.1
|
RP11-58C22.1
|
Uncharacterized protein |
| chr17_-_15466742 | 0.37 |
ENST00000584811.1
ENST00000419890.2 |
TVP23C
|
trans-golgi network vesicle protein 23 homolog C (S. cerevisiae) |
| chr13_-_47471155 | 0.37 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr2_+_110551851 | 0.36 |
ENST00000272454.6
ENST00000430736.1 ENST00000016946.3 ENST00000441344.1 |
RGPD5
|
RANBP2-like and GRIP domain containing 5 |
| chr6_-_30181133 | 0.35 |
ENST00000454678.2
ENST00000434785.1 |
TRIM26
|
tripartite motif containing 26 |
| chr9_-_19786926 | 0.35 |
ENST00000341998.2
ENST00000286344.3 |
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr8_+_104892639 | 0.35 |
ENST00000436393.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr3_+_173116225 | 0.35 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr14_+_39734482 | 0.35 |
ENST00000554392.1
ENST00000555716.1 ENST00000341749.3 ENST00000557038.1 |
CTAGE5
|
CTAGE family, member 5 |
| chr12_+_133758115 | 0.34 |
ENST00000541009.2
ENST00000592241.1 |
ZNF268
|
zinc finger protein 268 |
| chr19_-_55652290 | 0.34 |
ENST00000589745.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr1_-_39347255 | 0.34 |
ENST00000454994.2
ENST00000357771.3 |
GJA9
|
gap junction protein, alpha 9, 59kDa |
| chr6_-_30181156 | 0.34 |
ENST00000418026.1
ENST00000416596.1 ENST00000453195.1 |
TRIM26
|
tripartite motif containing 26 |
| chr11_+_71249071 | 0.34 |
ENST00000398534.3
|
KRTAP5-8
|
keratin associated protein 5-8 |
| chr18_+_616711 | 0.34 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr12_-_11287243 | 0.33 |
ENST00000539585.1
|
TAS2R30
|
taste receptor, type 2, member 30 |
| chr3_+_46395219 | 0.33 |
ENST00000445132.2
ENST00000292301.4 |
CCR2
|
chemokine (C-C motif) receptor 2 |
| chr5_-_75013193 | 0.32 |
ENST00000514838.2
ENST00000506164.1 ENST00000502826.1 ENST00000503835.1 ENST00000428202.2 ENST00000380475.2 |
POC5
|
POC5 centriolar protein |
| chr11_-_86383370 | 0.32 |
ENST00000526834.1
ENST00000359636.2 |
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr3_-_191000172 | 0.32 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr2_-_89417335 | 0.32 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr2_-_111334678 | 0.30 |
ENST00000329516.3
ENST00000330331.5 ENST00000446930.1 |
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr18_+_29671812 | 0.30 |
ENST00000261593.3
ENST00000578914.1 |
RNF138
|
ring finger protein 138, E3 ubiquitin protein ligase |
| chr11_-_1629693 | 0.30 |
ENST00000399685.1
|
KRTAP5-3
|
keratin associated protein 5-3 |
| chr1_-_205391178 | 0.29 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr17_+_18684563 | 0.29 |
ENST00000476139.1
|
TVP23B
|
trans-golgi network vesicle protein 23 homolog B (S. cerevisiae) |
| chr2_-_89266286 | 0.28 |
ENST00000464162.1
|
IGKV1-6
|
immunoglobulin kappa variable 1-6 |
| chr2_+_106468204 | 0.28 |
ENST00000425756.1
ENST00000393349.2 |
NCK2
|
NCK adaptor protein 2 |
| chr17_-_29641084 | 0.28 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr10_-_27443155 | 0.28 |
ENST00000427324.1
ENST00000326799.3 |
YME1L1
|
YME1-like 1 ATPase |
| chr16_-_72128270 | 0.27 |
ENST00000426362.2
|
TXNL4B
|
thioredoxin-like 4B |
| chr18_-_13915530 | 0.27 |
ENST00000327606.3
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr11_+_22688150 | 0.27 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr16_-_21868739 | 0.27 |
ENST00000415645.2
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr1_-_39395165 | 0.27 |
ENST00000372985.3
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr14_-_75518129 | 0.26 |
ENST00000556257.1
ENST00000557648.1 ENST00000553263.1 ENST00000355774.2 ENST00000380968.2 ENST00000238662.7 |
MLH3
|
mutL homolog 3 |
| chr17_-_71228357 | 0.26 |
ENST00000583024.1
ENST00000403627.3 ENST00000405159.3 ENST00000581110.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr18_-_51884204 | 0.26 |
ENST00000577499.1
ENST00000584040.1 ENST00000581310.1 |
STARD6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr2_+_234959323 | 0.26 |
ENST00000373368.1
ENST00000168148.3 |
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr2_-_217559517 | 0.26 |
ENST00000449583.1
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr12_-_39734783 | 0.26 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr15_-_58306295 | 0.26 |
ENST00000559517.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr7_-_22539771 | 0.25 |
ENST00000406890.2
ENST00000424363.1 |
STEAP1B
|
STEAP family member 1B |
| chr3_-_121379739 | 0.25 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr16_-_21868978 | 0.25 |
ENST00000357370.5
ENST00000451409.1 ENST00000341400.7 ENST00000518761.4 |
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr9_-_69229650 | 0.25 |
ENST00000416428.1
|
CBWD6
|
COBW domain containing 6 |
| chr4_-_57844989 | 0.25 |
ENST00000264230.4
|
NOA1
|
nitric oxide associated 1 |
| chr11_+_118175132 | 0.25 |
ENST00000361763.4
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex) |
| chr16_-_4817129 | 0.24 |
ENST00000545009.1
ENST00000219478.6 |
ZNF500
|
zinc finger protein 500 |
| chr2_-_109605663 | 0.24 |
ENST00000409271.1
ENST00000258443.2 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr18_+_616672 | 0.24 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr4_+_186990298 | 0.24 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr11_+_45825616 | 0.24 |
ENST00000442528.2
ENST00000456334.1 ENST00000526817.1 |
SLC35C1
|
solute carrier family 35 (GDP-fucose transporter), member C1 |
| chr4_+_175839506 | 0.24 |
ENST00000505141.1
ENST00000359240.3 ENST00000445694.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr16_-_29415350 | 0.24 |
ENST00000524087.1
|
NPIPB11
|
nuclear pore complex interacting protein family, member B11 |
| chr10_-_100995540 | 0.23 |
ENST00000370546.1
ENST00000404542.1 |
HPSE2
|
heparanase 2 |
| chr17_-_15466850 | 0.23 |
ENST00000438826.3
ENST00000225576.3 ENST00000519970.1 ENST00000518321.1 ENST00000428082.2 ENST00000522212.2 |
TVP23C
TVP23C-CDRT4
|
trans-golgi network vesicle protein 23 homolog C (S. cerevisiae) TVP23C-CDRT4 readthrough |
| chr16_-_46655538 | 0.23 |
ENST00000303383.3
|
SHCBP1
|
SHC SH2-domain binding protein 1 |
| chr15_-_49103235 | 0.22 |
ENST00000380950.2
|
CEP152
|
centrosomal protein 152kDa |
| chr10_+_118305435 | 0.22 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr20_+_58179582 | 0.22 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr1_-_23520755 | 0.22 |
ENST00000314113.3
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr9_-_86571628 | 0.22 |
ENST00000376344.3
|
C9orf64
|
chromosome 9 open reading frame 64 |
| chr2_+_190540711 | 0.22 |
ENST00000520309.1
|
ANKAR
|
ankyrin and armadillo repeat containing |
| chr4_-_68749745 | 0.22 |
ENST00000283916.6
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr17_+_71228740 | 0.22 |
ENST00000268942.8
ENST00000359042.2 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr19_-_53758094 | 0.21 |
ENST00000601828.1
ENST00000598513.1 ENST00000599012.1 ENST00000333952.4 ENST00000598806.1 |
ZNF677
|
zinc finger protein 677 |
| chr9_-_114246635 | 0.21 |
ENST00000338205.5
|
KIAA0368
|
KIAA0368 |
| chr1_-_247267580 | 0.21 |
ENST00000366501.1
ENST00000366500.1 ENST00000476158.2 ENST00000448299.2 ENST00000358785.4 ENST00000343381.6 |
ZNF669
|
zinc finger protein 669 |
| chr8_+_18248786 | 0.21 |
ENST00000520116.1
|
NAT2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr16_+_30935418 | 0.21 |
ENST00000338343.4
|
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr19_-_48614063 | 0.21 |
ENST00000599921.1
ENST00000599111.1 |
PLA2G4C
|
phospholipase A2, group IVC (cytosolic, calcium-independent) |
| chr1_-_23521222 | 0.21 |
ENST00000374619.1
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr4_+_57845043 | 0.21 |
ENST00000433463.1
ENST00000314595.5 |
POLR2B
|
polymerase (RNA) II (DNA directed) polypeptide B, 140kDa |
| chr18_-_64271363 | 0.21 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr14_+_37641012 | 0.21 |
ENST00000556667.1
|
SLC25A21-AS1
|
SLC25A21 antisense RNA 1 |
| chr8_+_18248755 | 0.21 |
ENST00000286479.3
|
NAT2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr1_+_156095951 | 0.20 |
ENST00000448611.2
ENST00000368297.1 |
LMNA
|
lamin A/C |
| chr4_+_70894130 | 0.20 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr8_+_32579341 | 0.20 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr19_-_48614033 | 0.20 |
ENST00000354276.3
|
PLA2G4C
|
phospholipase A2, group IVC (cytosolic, calcium-independent) |
| chr1_-_31661000 | 0.20 |
ENST00000263693.1
ENST00000398657.2 ENST00000526106.1 |
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr12_+_119772502 | 0.20 |
ENST00000536742.1
ENST00000327554.2 |
CCDC60
|
coiled-coil domain containing 60 |
| chr2_+_102413726 | 0.20 |
ENST00000350878.4
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr7_+_76090993 | 0.20 |
ENST00000425780.1
ENST00000456590.1 ENST00000451769.1 ENST00000324432.5 ENST00000307569.8 ENST00000457529.1 ENST00000446600.1 ENST00000413936.2 ENST00000423646.1 ENST00000438930.1 ENST00000430490.2 |
DTX2
|
deltex homolog 2 (Drosophila) |
| chr16_-_20753114 | 0.20 |
ENST00000396083.2
|
THUMPD1
|
THUMP domain containing 1 |
| chr10_+_5406935 | 0.19 |
ENST00000380433.3
|
UCN3
|
urocortin 3 |
| chr8_-_2585929 | 0.19 |
ENST00000519393.1
ENST00000520842.1 ENST00000520570.1 ENST00000517357.1 ENST00000517984.1 ENST00000523971.1 |
RP11-134O21.1
|
RP11-134O21.1 |
| chr21_+_33671264 | 0.19 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr9_-_139343294 | 0.19 |
ENST00000313084.5
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr12_+_32832134 | 0.19 |
ENST00000452533.2
|
DNM1L
|
dynamin 1-like |
| chrX_-_20159934 | 0.19 |
ENST00000379593.1
ENST00000379607.5 |
EIF1AX
|
eukaryotic translation initiation factor 1A, X-linked |
| chr1_+_211499957 | 0.18 |
ENST00000336184.2
|
TRAF5
|
TNF receptor-associated factor 5 |
| chr16_-_28374829 | 0.18 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family, member B6 |
| chr6_-_46703430 | 0.18 |
ENST00000537365.1
|
PLA2G7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr6_-_136847099 | 0.18 |
ENST00000438100.2
|
MAP7
|
microtubule-associated protein 7 |
| chr17_-_18430160 | 0.18 |
ENST00000392176.3
|
FAM106A
|
family with sequence similarity 106, member A |
| chr5_-_136834982 | 0.18 |
ENST00000510689.1
ENST00000394945.1 |
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
| chr11_-_86383650 | 0.18 |
ENST00000526944.1
ENST00000530335.1 ENST00000543262.1 ENST00000524826.1 |
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chrX_+_140982452 | 0.18 |
ENST00000544766.1
|
MAGEC3
|
melanoma antigen family C, 3 |
| chr3_+_36421826 | 0.18 |
ENST00000273183.3
|
STAC
|
SH3 and cysteine rich domain |
| chr19_-_12941469 | 0.18 |
ENST00000586969.1
ENST00000589681.1 ENST00000585384.1 ENST00000589808.1 |
RTBDN
|
retbindin |
| chr7_-_3214287 | 0.18 |
ENST00000404626.3
|
AC091801.1
|
LOC392621; Uncharacterized protein |
| chr6_-_15663198 | 0.17 |
ENST00000338950.5
ENST00000511762.2 ENST00000355917.3 ENST00000344537.5 |
DTNBP1
|
dystrobrevin binding protein 1 |
| chr13_-_79233314 | 0.17 |
ENST00000282003.6
|
RNF219
|
ring finger protein 219 |
| chr3_-_139396560 | 0.17 |
ENST00000514703.1
ENST00000511444.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr1_+_53308398 | 0.17 |
ENST00000371528.1
|
ZYG11A
|
zyg-11 family member A, cell cycle regulator |
| chr2_+_190541153 | 0.17 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chr6_-_116381918 | 0.17 |
ENST00000606080.1
|
FRK
|
fyn-related kinase |
| chr9_-_117420052 | 0.17 |
ENST00000423632.1
|
RP11-402G3.5
|
RP11-402G3.5 |
| chr1_+_196743912 | 0.17 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr6_+_42749759 | 0.16 |
ENST00000314073.5
|
GLTSCR1L
|
GLTSCR1-like |
| chr6_+_121756809 | 0.16 |
ENST00000282561.3
|
GJA1
|
gap junction protein, alpha 1, 43kDa |
| chr8_+_85095553 | 0.16 |
ENST00000521268.1
|
RALYL
|
RALY RNA binding protein-like |
| chr7_-_124569991 | 0.16 |
ENST00000446993.1
ENST00000357628.3 ENST00000393329.1 |
POT1
|
protection of telomeres 1 |
| chr7_-_38305279 | 0.16 |
ENST00000443402.2
|
TRGC1
|
T cell receptor gamma constant 1 |
| chr16_+_30064444 | 0.16 |
ENST00000395248.1
ENST00000566897.1 ENST00000568435.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr1_+_196743943 | 0.16 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr12_-_28125638 | 0.15 |
ENST00000545234.1
|
PTHLH
|
parathyroid hormone-like hormone |
| chr8_+_67624653 | 0.15 |
ENST00000521198.2
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr13_+_98612446 | 0.15 |
ENST00000496368.1
ENST00000421861.2 ENST00000357602.3 |
IPO5
|
importin 5 |
| chr8_+_93895865 | 0.15 |
ENST00000391681.1
|
AC117834.1
|
AC117834.1 |
| chr1_-_72566613 | 0.15 |
ENST00000306821.3
|
NEGR1
|
neuronal growth regulator 1 |
| chr12_-_53601055 | 0.15 |
ENST00000552972.1
ENST00000422257.3 ENST00000267082.5 |
ITGB7
|
integrin, beta 7 |
| chr16_-_11375179 | 0.15 |
ENST00000312511.3
|
PRM1
|
protamine 1 |
| chr16_+_56970567 | 0.15 |
ENST00000563911.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr1_-_145382362 | 0.15 |
ENST00000419817.1
ENST00000421937.3 ENST00000433081.2 |
RP11-458D21.1
|
RP11-458D21.1 |
| chr22_+_24198890 | 0.15 |
ENST00000345044.6
|
SLC2A11
|
solute carrier family 2 (facilitated glucose transporter), member 11 |
| chr4_+_15341442 | 0.15 |
ENST00000397700.2
ENST00000295297.4 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr3_-_93747425 | 0.15 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr6_+_53948328 | 0.15 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr16_+_30064411 | 0.14 |
ENST00000338110.5
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr14_-_106725723 | 0.14 |
ENST00000390609.2
|
IGHV3-23
|
immunoglobulin heavy variable 3-23 |
| chr8_+_85095769 | 0.14 |
ENST00000518566.1
|
RALYL
|
RALY RNA binding protein-like |
| chr14_-_53162361 | 0.14 |
ENST00000395686.3
|
ERO1L
|
ERO1-like (S. cerevisiae) |
| chr12_+_82347498 | 0.14 |
ENST00000550506.1
|
RP11-362A1.1
|
RP11-362A1.1 |
| chrX_+_1387693 | 0.14 |
ENST00000381529.3
ENST00000432318.2 ENST00000361536.3 ENST00000501036.2 ENST00000381524.3 ENST00000412290.1 |
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr5_+_68513622 | 0.14 |
ENST00000512880.1
ENST00000602380.1 |
MRPS36
|
mitochondrial ribosomal protein S36 |
| chr8_+_39759794 | 0.14 |
ENST00000518804.1
ENST00000519154.1 ENST00000522495.1 ENST00000522840.1 |
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr14_+_22694606 | 0.14 |
ENST00000390463.3
|
TRAV36DV7
|
T cell receptor alpha variable 36/delta variable 7 |
| chr2_-_70418032 | 0.14 |
ENST00000425268.1
ENST00000428751.1 ENST00000417203.1 ENST00000417865.1 ENST00000428010.1 ENST00000447804.1 ENST00000264434.2 |
C2orf42
|
chromosome 2 open reading frame 42 |
| chr5_+_96038476 | 0.14 |
ENST00000511049.1
ENST00000309190.5 ENST00000510156.1 ENST00000509903.1 ENST00000511782.1 ENST00000504465.1 |
CAST
|
calpastatin |
| chr7_+_64838786 | 0.14 |
ENST00000450302.2
|
ZNF92
|
zinc finger protein 92 |
| chr12_+_32832203 | 0.13 |
ENST00000553257.1
ENST00000549701.1 ENST00000358214.5 ENST00000266481.6 ENST00000551476.1 ENST00000550154.1 ENST00000547312.1 ENST00000414834.2 ENST00000381000.4 ENST00000548750.1 |
DNM1L
|
dynamin 1-like |
| chr5_-_10308125 | 0.13 |
ENST00000296658.3
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr5_+_141346385 | 0.13 |
ENST00000513019.1
ENST00000356143.1 |
RNF14
|
ring finger protein 14 |
| chr19_+_49199209 | 0.13 |
ENST00000522966.1
ENST00000425340.2 ENST00000391876.4 |
FUT2
|
fucosyltransferase 2 (secretor status included) |
| chr10_-_69597828 | 0.13 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr14_-_101295407 | 0.13 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr16_-_30621663 | 0.13 |
ENST00000287461.3
|
ZNF689
|
zinc finger protein 689 |
| chr3_+_118892362 | 0.13 |
ENST00000497685.1
ENST00000264234.3 |
UPK1B
|
uroplakin 1B |
| chr3_+_39424828 | 0.13 |
ENST00000273158.4
ENST00000431510.1 |
SLC25A38
|
solute carrier family 25, member 38 |
| chr14_-_69261310 | 0.12 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chrX_-_15333736 | 0.12 |
ENST00000380470.3
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr9_-_21482312 | 0.12 |
ENST00000448696.3
|
IFNE
|
interferon, epsilon |
| chr19_+_3762645 | 0.12 |
ENST00000330133.4
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr10_+_97733786 | 0.12 |
ENST00000371198.2
|
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr10_-_48806939 | 0.12 |
ENST00000374233.3
ENST00000507417.1 ENST00000512321.1 ENST00000395660.2 ENST00000374235.2 ENST00000395661.3 |
PTPN20B
|
protein tyrosine phosphatase, non-receptor type 20B |
| chr12_+_124457746 | 0.12 |
ENST00000392404.3
ENST00000538932.2 ENST00000337815.4 ENST00000540762.2 |
ZNF664
FAM101A
|
zinc finger protein 664 family with sequence similarity 101, member A |
| chr8_+_85095497 | 0.12 |
ENST00000522455.1
ENST00000521695.1 |
RALYL
|
RALY RNA binding protein-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID3A
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.2 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.3 | 0.8 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.2 | 0.8 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.2 | 1.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 0.5 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.4 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 0.1 | 0.7 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.1 | 0.5 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.3 | GO:1902567 | negative regulation of eosinophil activation(GO:1902567) |
| 0.1 | 0.3 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.1 | 0.6 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 1.7 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.1 | 0.4 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.6 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.5 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.3 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 | 0.8 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 0.4 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.2 | GO:0045356 | microglial cell activation involved in immune response(GO:0002282) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 0.2 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 | 0.5 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.1 | 0.2 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 | 0.2 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.2 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.3 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.3 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.5 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.4 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.0 | 0.5 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.6 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.3 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.3 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 2.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.2 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.1 | GO:0098758 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.0 | 0.5 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.2 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.7 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.2 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.8 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.3 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) |
| 0.0 | 0.3 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 1.2 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 2.3 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 1.0 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.2 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.4 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.2 | GO:0032202 | telomere assembly(GO:0032202) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.2 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.0 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.4 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.5 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.3 | GO:0005712 | chiasma(GO:0005712) |
| 0.1 | 0.3 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 0.5 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.5 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.7 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 0.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 2.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 2.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 0.5 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.2 | 0.7 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.1 | 0.4 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.1 | 0.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.4 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.5 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.8 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 0.2 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.4 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.1 | 0.6 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.2 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 0.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.2 | GO:0016160 | amylase activity(GO:0016160) |
| 0.0 | 0.2 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.6 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.0 | 0.5 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.3 | GO:0019237 | satellite DNA binding(GO:0003696) centromeric DNA binding(GO:0019237) |
| 0.0 | 0.3 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 1.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.6 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.5 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.5 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 4.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 0.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 2.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 3.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |