Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for ARID5A
Z-value: 0.52
Transcription factors associated with ARID5A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID5A
|
ENSG00000196843.11 | AT-rich interaction domain 5A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID5A | hg19_v2_chr2_+_97203082_97203159, hg19_v2_chr2_+_97202480_97202499 | 0.12 | 5.6e-01 | Click! |
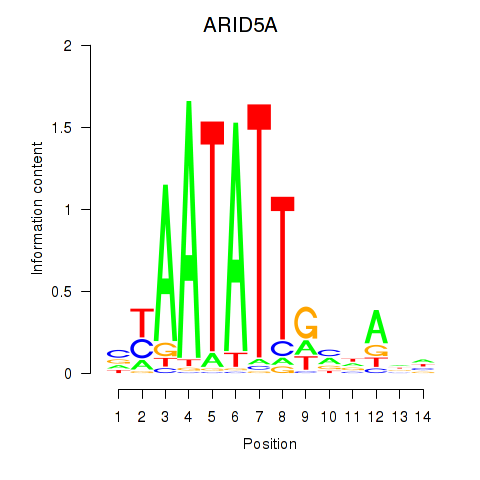
Activity profile of ARID5A motif
Sorted Z-values of ARID5A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_21207503 | 2.85 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr3_-_74570291 | 0.81 |
ENST00000263665.6
|
CNTN3
|
contactin 3 (plasmacytoma associated) |
| chr3_-_141747950 | 0.68 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr12_+_21284118 | 0.48 |
ENST00000256958.2
|
SLCO1B1
|
solute carrier organic anion transporter family, member 1B1 |
| chr1_-_169703203 | 0.47 |
ENST00000333360.7
ENST00000367781.4 ENST00000367782.4 ENST00000367780.4 ENST00000367779.4 |
SELE
|
selectin E |
| chr11_+_5474638 | 0.34 |
ENST00000341449.2
|
OR51I2
|
olfactory receptor, family 51, subfamily I, member 2 |
| chr4_-_177116772 | 0.34 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr3_-_108672742 | 0.34 |
ENST00000261047.3
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr3_+_101546827 | 0.32 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr11_-_102595681 | 0.30 |
ENST00000236826.3
|
MMP8
|
matrix metallopeptidase 8 (neutrophil collagenase) |
| chr16_-_4665023 | 0.30 |
ENST00000591897.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr16_-_4664860 | 0.29 |
ENST00000587615.1
ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1
|
UBA-like domain containing 1 |
| chr12_+_9980069 | 0.29 |
ENST00000354855.3
ENST00000324214.4 ENST00000279544.3 |
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr4_-_69111401 | 0.27 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr1_-_145589424 | 0.26 |
ENST00000334513.5
|
NUDT17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr3_+_190333097 | 0.26 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr11_+_34664014 | 0.24 |
ENST00000527935.1
|
EHF
|
ets homologous factor |
| chr6_-_25874440 | 0.24 |
ENST00000361703.6
ENST00000397060.4 |
SLC17A3
|
solute carrier family 17 (organic anion transporter), member 3 |
| chr11_+_74459876 | 0.23 |
ENST00000299563.4
|
RNF169
|
ring finger protein 169 |
| chr9_-_3469181 | 0.22 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr3_+_97868170 | 0.21 |
ENST00000437310.1
|
OR5H14
|
olfactory receptor, family 5, subfamily H, member 14 |
| chr10_+_127661942 | 0.21 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr2_-_89545079 | 0.21 |
ENST00000468494.1
|
IGKV2-30
|
immunoglobulin kappa variable 2-30 |
| chr22_-_38699003 | 0.21 |
ENST00000451964.1
|
CSNK1E
|
casein kinase 1, epsilon |
| chr3_+_148447887 | 0.21 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr11_+_4788500 | 0.20 |
ENST00000380390.1
|
MMP26
|
matrix metallopeptidase 26 |
| chr9_-_140196703 | 0.20 |
ENST00000356628.2
|
NRARP
|
NOTCH-regulated ankyrin repeat protein |
| chr1_+_116654376 | 0.20 |
ENST00000369500.3
|
MAB21L3
|
mab-21-like 3 (C. elegans) |
| chr1_+_168250194 | 0.19 |
ENST00000367821.3
|
TBX19
|
T-box 19 |
| chr7_+_13141097 | 0.19 |
ENST00000411542.1
|
AC011288.2
|
AC011288.2 |
| chr18_+_61223393 | 0.19 |
ENST00000269491.1
ENST00000382768.1 |
SERPINB12
|
serpin peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr6_-_26056695 | 0.19 |
ENST00000343677.2
|
HIST1H1C
|
histone cluster 1, H1c |
| chr7_-_7782204 | 0.19 |
ENST00000418534.2
|
AC007161.5
|
AC007161.5 |
| chr18_+_29171689 | 0.19 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr11_+_73661364 | 0.18 |
ENST00000339764.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr13_-_41706864 | 0.18 |
ENST00000379485.1
ENST00000499385.2 |
KBTBD6
|
kelch repeat and BTB (POZ) domain containing 6 |
| chr5_-_86534822 | 0.17 |
ENST00000445770.2
|
AC008394.1
|
Uncharacterized protein |
| chr12_-_11062161 | 0.17 |
ENST00000390677.2
|
TAS2R13
|
taste receptor, type 2, member 13 |
| chr14_+_56584414 | 0.17 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr11_-_114466477 | 0.17 |
ENST00000375478.3
|
NXPE4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr1_+_196912902 | 0.17 |
ENST00000476712.2
ENST00000367415.5 |
CFHR2
|
complement factor H-related 2 |
| chr2_-_69180083 | 0.17 |
ENST00000328895.4
|
GKN2
|
gastrokine 2 |
| chr3_-_108672609 | 0.17 |
ENST00000393963.3
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr1_+_67632083 | 0.16 |
ENST00000347310.5
ENST00000371002.1 |
IL23R
|
interleukin 23 receptor |
| chr12_+_10460549 | 0.16 |
ENST00000543420.1
ENST00000543777.1 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr12_+_12938541 | 0.16 |
ENST00000356591.4
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr21_+_43823983 | 0.16 |
ENST00000291535.6
ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr12_+_131438443 | 0.16 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chr3_+_94657086 | 0.15 |
ENST00000463200.1
|
LINC00879
|
long intergenic non-protein coding RNA 879 |
| chr17_+_68071458 | 0.15 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr21_-_35899113 | 0.15 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr5_+_159848807 | 0.15 |
ENST00000352433.5
|
PTTG1
|
pituitary tumor-transforming 1 |
| chr16_-_4588822 | 0.15 |
ENST00000564828.1
|
CDIP1
|
cell death-inducing p53 target 1 |
| chr1_-_149459549 | 0.15 |
ENST00000369175.3
|
FAM72C
|
family with sequence similarity 72, member C |
| chr12_-_96390063 | 0.15 |
ENST00000541929.1
|
HAL
|
histidine ammonia-lyase |
| chr12_+_48099858 | 0.15 |
ENST00000547799.1
|
RP1-197B17.3
|
RP1-197B17.3 |
| chr6_+_132455118 | 0.15 |
ENST00000458028.1
|
LINC01013
|
long intergenic non-protein coding RNA 1013 |
| chr5_+_159848854 | 0.15 |
ENST00000517480.1
ENST00000520452.1 ENST00000393964.1 |
PTTG1
|
pituitary tumor-transforming 1 |
| chr9_+_35906176 | 0.14 |
ENST00000354323.2
|
HRCT1
|
histidine rich carboxyl terminus 1 |
| chr11_+_55650773 | 0.14 |
ENST00000449290.2
|
TRIM51
|
tripartite motif-containing 51 |
| chr4_-_47983519 | 0.14 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr2_+_12246664 | 0.14 |
ENST00000449986.1
|
AC096559.1
|
AC096559.1 |
| chr4_+_6271558 | 0.14 |
ENST00000503569.1
ENST00000226760.1 |
WFS1
|
Wolfram syndrome 1 (wolframin) |
| chr12_+_8995832 | 0.14 |
ENST00000541459.1
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chrX_+_55246771 | 0.14 |
ENST00000289619.5
ENST00000374955.3 |
PAGE5
|
P antigen family, member 5 (prostate associated) |
| chrX_+_36053908 | 0.14 |
ENST00000378660.2
|
CHDC2
|
calponin homology domain containing 2 |
| chr7_+_39125365 | 0.14 |
ENST00000559001.1
ENST00000464276.2 |
POU6F2
|
POU class 6 homeobox 2 |
| chr1_+_77333117 | 0.14 |
ENST00000477717.1
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr1_+_173793777 | 0.14 |
ENST00000239457.5
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr3_-_18466026 | 0.14 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr20_+_55904815 | 0.14 |
ENST00000371263.3
ENST00000345868.4 ENST00000371260.4 ENST00000418127.1 |
SPO11
|
SPO11 meiotic protein covalently bound to DSB |
| chr16_-_4588762 | 0.13 |
ENST00000562334.1
ENST00000562579.1 ENST00000567695.1 ENST00000563507.1 |
CDIP1
|
cell death-inducing p53 target 1 |
| chr8_-_57472154 | 0.13 |
ENST00000499425.1
ENST00000523664.1 ENST00000518943.1 ENST00000524338.1 |
LINC00968
|
long intergenic non-protein coding RNA 968 |
| chr12_-_91546926 | 0.13 |
ENST00000550758.1
|
DCN
|
decorin |
| chrX_-_74376108 | 0.13 |
ENST00000339447.4
ENST00000373394.3 ENST00000529949.1 ENST00000534524.1 ENST00000253577.3 |
ABCB7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr3_-_87325612 | 0.13 |
ENST00000561167.1
ENST00000560656.1 ENST00000344265.3 |
POU1F1
|
POU class 1 homeobox 1 |
| chr1_+_117963209 | 0.13 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr13_+_43355732 | 0.13 |
ENST00000313851.1
|
FAM216B
|
family with sequence similarity 216, member B |
| chr1_-_197036364 | 0.13 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr10_+_78078088 | 0.13 |
ENST00000496424.2
|
C10orf11
|
chromosome 10 open reading frame 11 |
| chr13_+_43355683 | 0.12 |
ENST00000537894.1
|
FAM216B
|
family with sequence similarity 216, member B |
| chr1_+_158815588 | 0.12 |
ENST00000438394.1
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr4_-_143226979 | 0.12 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr20_-_5426332 | 0.12 |
ENST00000420529.1
|
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chr6_+_24357131 | 0.12 |
ENST00000274766.1
|
KAAG1
|
kidney associated antigen 1 |
| chr16_-_31439735 | 0.12 |
ENST00000287490.4
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr10_+_5005598 | 0.12 |
ENST00000442997.1
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr18_+_61554932 | 0.12 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr6_+_30035307 | 0.11 |
ENST00000376765.2
ENST00000376763.1 |
PPP1R11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr8_+_36641842 | 0.11 |
ENST00000523973.1
ENST00000399881.3 |
KCNU1
|
potassium channel, subfamily U, member 1 |
| chr6_+_26156551 | 0.11 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr3_-_87325728 | 0.11 |
ENST00000350375.2
|
POU1F1
|
POU class 1 homeobox 1 |
| chr2_+_201754050 | 0.11 |
ENST00000426253.1
ENST00000416651.1 ENST00000454952.1 ENST00000409020.1 ENST00000359683.4 |
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr15_-_72523924 | 0.11 |
ENST00000566809.1
ENST00000567087.1 ENST00000569050.1 ENST00000568883.1 |
PKM
|
pyruvate kinase, muscle |
| chr3_+_16306837 | 0.11 |
ENST00000606098.1
|
OXNAD1
|
oxidoreductase NAD-binding domain containing 1 |
| chr19_+_35849362 | 0.11 |
ENST00000327809.4
|
FFAR3
|
free fatty acid receptor 3 |
| chr10_+_134150835 | 0.11 |
ENST00000432555.2
|
LRRC27
|
leucine rich repeat containing 27 |
| chr6_-_49834209 | 0.11 |
ENST00000507853.1
|
CRISP1
|
cysteine-rich secretory protein 1 |
| chr2_+_201754135 | 0.11 |
ENST00000409357.1
ENST00000409129.2 |
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr9_+_116207007 | 0.11 |
ENST00000374140.2
|
RGS3
|
regulator of G-protein signaling 3 |
| chr21_+_17553910 | 0.11 |
ENST00000428669.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr13_+_75126978 | 0.11 |
ENST00000596240.1
ENST00000451336.2 |
LINC00347
|
long intergenic non-protein coding RNA 347 |
| chr6_+_46761118 | 0.11 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr9_-_34381511 | 0.11 |
ENST00000379124.1
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr6_-_49834240 | 0.10 |
ENST00000335847.4
|
CRISP1
|
cysteine-rich secretory protein 1 |
| chr1_-_165668100 | 0.10 |
ENST00000354775.4
|
ALDH9A1
|
aldehyde dehydrogenase 9 family, member A1 |
| chr6_+_123317116 | 0.10 |
ENST00000275162.5
|
CLVS2
|
clavesin 2 |
| chr6_-_76782371 | 0.10 |
ENST00000369950.3
ENST00000369963.3 |
IMPG1
|
interphotoreceptor matrix proteoglycan 1 |
| chr6_-_130543958 | 0.10 |
ENST00000437477.2
ENST00000439090.2 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr9_+_77230499 | 0.10 |
ENST00000396204.2
|
RORB
|
RAR-related orphan receptor B |
| chrX_+_10031499 | 0.10 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr1_+_47489240 | 0.10 |
ENST00000371901.3
|
CYP4X1
|
cytochrome P450, family 4, subfamily X, polypeptide 1 |
| chr18_-_50240 | 0.10 |
ENST00000573909.1
|
RP11-683L23.1
|
Tubulin beta-8 chain-like protein LOC260334 |
| chr5_+_54398463 | 0.10 |
ENST00000274306.6
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
| chr5_-_55412774 | 0.10 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr2_-_232395169 | 0.10 |
ENST00000305141.4
|
NMUR1
|
neuromedin U receptor 1 |
| chr4_-_143227088 | 0.09 |
ENST00000511838.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr1_+_196788887 | 0.09 |
ENST00000320493.5
ENST00000367424.4 ENST00000367421.3 |
CFHR1
CFHR2
|
complement factor H-related 1 complement factor H-related 2 |
| chrX_+_79675965 | 0.09 |
ENST00000308293.5
|
FAM46D
|
family with sequence similarity 46, member D |
| chr20_+_15177480 | 0.09 |
ENST00000402914.1
|
MACROD2
|
MACRO domain containing 2 |
| chr1_-_203320617 | 0.09 |
ENST00000354955.4
|
FMOD
|
fibromodulin |
| chr10_-_127505167 | 0.09 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr3_-_112320749 | 0.09 |
ENST00000610103.1
|
RP11-572C15.6
|
RP11-572C15.6 |
| chr17_+_38975358 | 0.09 |
ENST00000436612.1
ENST00000301665.3 |
TMEM99
|
transmembrane protein 99 |
| chr9_-_34381536 | 0.09 |
ENST00000379126.3
ENST00000379127.1 ENST00000379133.3 |
C9orf24
|
chromosome 9 open reading frame 24 |
| chr6_-_15548591 | 0.09 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr6_-_26199471 | 0.09 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr2_-_180726232 | 0.09 |
ENST00000410066.1
|
ZNF385B
|
zinc finger protein 385B |
| chr1_-_157014865 | 0.09 |
ENST00000361409.2
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr20_+_30102231 | 0.09 |
ENST00000335574.5
ENST00000340852.5 ENST00000398174.3 ENST00000376127.3 ENST00000344042.5 |
HM13
|
histocompatibility (minor) 13 |
| chr3_-_107596910 | 0.09 |
ENST00000464359.2
ENST00000464823.1 ENST00000466155.1 ENST00000473528.2 ENST00000608306.1 ENST00000488852.1 ENST00000608137.1 ENST00000608307.1 ENST00000609429.1 ENST00000601385.1 ENST00000475362.1 ENST00000600240.1 ENST00000600749.1 |
LINC00635
|
long intergenic non-protein coding RNA 635 |
| chr5_-_35089722 | 0.09 |
ENST00000511486.1
ENST00000310101.5 ENST00000231423.3 ENST00000513753.1 ENST00000348262.3 ENST00000397391.3 ENST00000542609.1 |
PRLR
|
prolactin receptor |
| chr6_-_26235206 | 0.09 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr5_+_57787254 | 0.08 |
ENST00000502276.1
ENST00000396776.2 ENST00000511930.1 |
GAPT
|
GRB2-binding adaptor protein, transmembrane |
| chr1_+_74701062 | 0.08 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chrX_+_135614293 | 0.08 |
ENST00000370634.3
|
VGLL1
|
vestigial like 1 (Drosophila) |
| chr1_+_40713573 | 0.08 |
ENST00000372766.3
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr6_+_33043703 | 0.08 |
ENST00000418931.2
ENST00000535465.1 |
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr17_+_6554971 | 0.08 |
ENST00000391428.2
|
C17orf100
|
chromosome 17 open reading frame 100 |
| chr18_+_61575200 | 0.08 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr19_+_55014085 | 0.08 |
ENST00000351841.2
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2 |
| chrX_+_22050546 | 0.08 |
ENST00000379374.4
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr15_+_69307028 | 0.08 |
ENST00000388866.3
ENST00000530406.2 |
NOX5
|
NADPH oxidase, EF-hand calcium binding domain 5 |
| chr15_-_54025300 | 0.08 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr1_-_46642154 | 0.08 |
ENST00000540385.1
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr12_-_96390108 | 0.08 |
ENST00000538703.1
ENST00000261208.3 |
HAL
|
histidine ammonia-lyase |
| chr14_-_45252031 | 0.08 |
ENST00000556405.1
|
RP11-398E10.1
|
RP11-398E10.1 |
| chr4_+_55095428 | 0.08 |
ENST00000508170.1
ENST00000512143.1 |
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr2_+_210444748 | 0.08 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr16_+_15528332 | 0.08 |
ENST00000566490.1
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr4_+_119199904 | 0.08 |
ENST00000602483.1
ENST00000602819.1 |
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr4_-_155511887 | 0.08 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr1_+_948803 | 0.08 |
ENST00000379389.4
|
ISG15
|
ISG15 ubiquitin-like modifier |
| chr3_+_148415571 | 0.08 |
ENST00000497524.1
ENST00000349243.3 ENST00000542281.1 ENST00000418473.2 ENST00000404754.2 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr10_+_114710516 | 0.08 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr1_-_158656488 | 0.08 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr12_-_8218997 | 0.08 |
ENST00000307637.4
|
C3AR1
|
complement component 3a receptor 1 |
| chr20_-_1309809 | 0.07 |
ENST00000360779.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr8_-_124749609 | 0.07 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr9_-_118687405 | 0.07 |
ENST00000374014.3
|
LINC00474
|
long intergenic non-protein coding RNA 474 |
| chr11_+_60145997 | 0.07 |
ENST00000530614.1
ENST00000530027.1 ENST00000530234.2 ENST00000528215.1 ENST00000531787.1 |
MS4A7
MS4A14
|
membrane-spanning 4-domains, subfamily A, member 7 membrane-spanning 4-domains, subfamily A, member 14 |
| chr1_+_76540386 | 0.07 |
ENST00000328299.3
|
ST6GALNAC3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr3_-_167191814 | 0.07 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr1_-_89736434 | 0.07 |
ENST00000370459.3
|
GBP5
|
guanylate binding protein 5 |
| chr19_+_56368803 | 0.07 |
ENST00000587891.1
|
NLRP4
|
NLR family, pyrin domain containing 4 |
| chrX_+_36065053 | 0.07 |
ENST00000313548.4
|
CHDC2
|
calponin homology domain containing 2 |
| chr6_-_33239712 | 0.07 |
ENST00000436044.2
|
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr6_-_52859046 | 0.07 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr2_+_219646462 | 0.07 |
ENST00000258415.4
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1 |
| chr7_-_142120321 | 0.07 |
ENST00000390377.1
|
TRBV7-7
|
T cell receptor beta variable 7-7 |
| chr12_+_4758264 | 0.07 |
ENST00000266544.5
|
NDUFA9
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa |
| chr2_-_105030466 | 0.07 |
ENST00000449772.1
|
AC068535.3
|
AC068535.3 |
| chr19_+_55014013 | 0.07 |
ENST00000301202.2
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2 |
| chr7_-_19748640 | 0.07 |
ENST00000222567.5
|
TWISTNB
|
TWIST neighbor |
| chr6_+_132455526 | 0.07 |
ENST00000443303.1
|
LINC01013
|
long intergenic non-protein coding RNA 1013 |
| chr4_+_55095264 | 0.07 |
ENST00000257290.5
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr3_+_101280677 | 0.07 |
ENST00000309922.6
ENST00000495642.1 |
TRMT10C
|
tRNA methyltransferase 10 homolog C (S. cerevisiae) |
| chr7_+_148892557 | 0.07 |
ENST00000262085.3
|
ZNF282
|
zinc finger protein 282 |
| chr11_-_114466471 | 0.07 |
ENST00000424261.2
|
NXPE4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr14_-_60952739 | 0.07 |
ENST00000555476.1
ENST00000321731.3 |
C14orf39
|
chromosome 14 open reading frame 39 |
| chr6_+_88106840 | 0.07 |
ENST00000369570.4
|
C6orf164
|
chromosome 6 open reading frame 164 |
| chr1_+_160336851 | 0.07 |
ENST00000302101.5
|
NHLH1
|
nescient helix loop helix 1 |
| chr10_-_28623368 | 0.07 |
ENST00000441595.2
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr22_-_29457832 | 0.07 |
ENST00000216071.4
|
C22orf31
|
chromosome 22 open reading frame 31 |
| chr4_+_70796784 | 0.07 |
ENST00000246891.4
ENST00000444405.3 |
CSN1S1
|
casein alpha s1 |
| chr4_+_119199864 | 0.07 |
ENST00000602414.1
ENST00000602520.1 |
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr14_-_25045446 | 0.07 |
ENST00000216336.2
|
CTSG
|
cathepsin G |
| chrX_+_36246735 | 0.07 |
ENST00000378653.3
|
CXorf30
|
chromosome X open reading frame 30 |
| chr11_+_55029628 | 0.07 |
ENST00000417545.2
|
TRIM48
|
tripartite motif containing 48 |
| chrX_+_8432871 | 0.07 |
ENST00000381032.1
ENST00000453306.1 ENST00000444481.1 |
VCX3B
|
variable charge, X-linked 3B |
| chr1_+_15256230 | 0.06 |
ENST00000376028.4
ENST00000400798.2 |
KAZN
|
kazrin, periplakin interacting protein |
| chrX_+_107683096 | 0.06 |
ENST00000328300.6
ENST00000361603.2 |
COL4A5
|
collagen, type IV, alpha 5 |
| chr1_+_196946680 | 0.06 |
ENST00000256785.4
|
CFHR5
|
complement factor H-related 5 |
| chr10_-_126716459 | 0.06 |
ENST00000309035.6
|
CTBP2
|
C-terminal binding protein 2 |
| chr12_+_58087901 | 0.06 |
ENST00000315970.7
ENST00000547079.1 ENST00000439210.2 ENST00000389146.6 ENST00000413095.2 ENST00000551035.1 ENST00000257966.8 ENST00000435406.2 ENST00000550372.1 ENST00000389142.5 |
OS9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr3_-_170744498 | 0.06 |
ENST00000382808.4
ENST00000314251.3 |
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr10_+_96443378 | 0.06 |
ENST00000285979.6
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18 |
| chr9_-_138391692 | 0.06 |
ENST00000429260.2
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chrX_-_18690210 | 0.06 |
ENST00000379984.3
|
RS1
|
retinoschisin 1 |
| chr7_-_129691201 | 0.06 |
ENST00000480193.1
ENST00000360708.5 ENST00000311873.5 ENST00000481503.1 ENST00000358303.4 |
ZC3HC1
|
zinc finger, C3HC-type containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID5A
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.3 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 3.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.2 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.2 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break processing(GO:0000706) meiotic DNA double-strand break formation(GO:0042138) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.4 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.3 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.5 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.1 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.3 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.5 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.0 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.2 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.1 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.0 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0070943 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) neutrophil mediated killing of bacterium(GO:0070944) |
| 0.0 | 0.1 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.0 | GO:1900276 | negative regulation of phospholipase C activity(GO:1900275) regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.2 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.0 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.4 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.0 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.5 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.3 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.2 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.2 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.3 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0016429 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.0 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.0 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |