Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
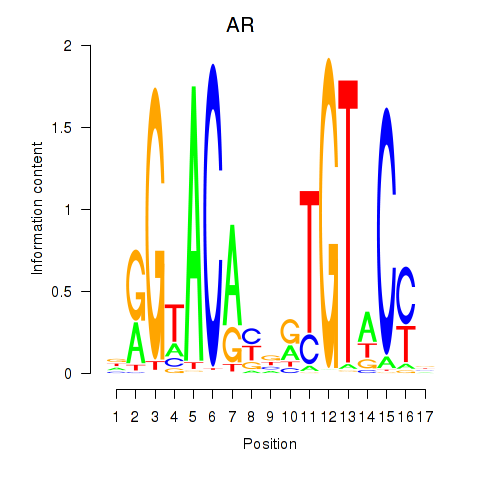
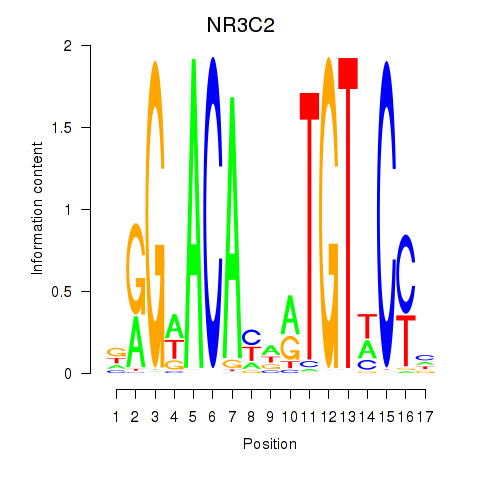
Results for AR_NR3C2
Z-value: 0.45
Transcription factors associated with AR_NR3C2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
AR
|
ENSG00000169083.11 | androgen receptor |
|
NR3C2
|
ENSG00000151623.10 | nuclear receptor subfamily 3 group C member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| AR | hg19_v2_chrX_+_66764375_66764465 | -0.16 | 4.4e-01 | Click! |
| NR3C2 | hg19_v2_chr4_-_149363662_149363688 | -0.02 | 9.3e-01 | Click! |
Activity profile of AR_NR3C2 motif
Sorted Z-values of AR_NR3C2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_56642041 | 0.97 |
ENST00000245185.5
|
MT2A
|
metallothionein 2A |
| chr16_+_56666563 | 0.86 |
ENST00000570233.1
|
MT1M
|
metallothionein 1M |
| chr16_+_56642489 | 0.86 |
ENST00000561491.1
|
MT2A
|
metallothionein 2A |
| chr16_+_56659687 | 0.84 |
ENST00000568293.1
ENST00000330439.6 |
MT1E
|
metallothionein 1E |
| chr2_+_102953608 | 0.80 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr16_+_56716336 | 0.77 |
ENST00000394485.4
ENST00000562939.1 |
MT1X
|
metallothionein 1X |
| chr22_+_30477000 | 0.76 |
ENST00000403975.1
|
HORMAD2
|
HORMA domain containing 2 |
| chr6_+_127898312 | 0.66 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr3_-_58563094 | 0.61 |
ENST00000464064.1
|
FAM107A
|
family with sequence similarity 107, member A |
| chr1_-_1149506 | 0.56 |
ENST00000379236.3
|
TNFRSF4
|
tumor necrosis factor receptor superfamily, member 4 |
| chr19_+_8117636 | 0.53 |
ENST00000253451.4
ENST00000315626.4 |
CCL25
|
chemokine (C-C motif) ligand 25 |
| chr19_+_35630628 | 0.49 |
ENST00000588715.1
ENST00000588607.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr19_+_35630926 | 0.48 |
ENST00000588081.1
ENST00000589121.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr19_-_45657028 | 0.43 |
ENST00000429338.1
ENST00000589776.1 |
NKPD1
|
NTPase, KAP family P-loop domain containing 1 |
| chr19_+_35630344 | 0.42 |
ENST00000455515.2
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chrX_+_8432871 | 0.41 |
ENST00000381032.1
ENST00000453306.1 ENST00000444481.1 |
VCX3B
|
variable charge, X-linked 3B |
| chr10_+_60759378 | 0.40 |
ENST00000432535.1
|
LINC00844
|
long intergenic non-protein coding RNA 844 |
| chr19_+_8117881 | 0.37 |
ENST00000390669.3
|
CCL25
|
chemokine (C-C motif) ligand 25 |
| chrX_-_6453159 | 0.37 |
ENST00000381089.3
ENST00000398729.1 |
VCX3A
|
variable charge, X-linked 3A |
| chr12_+_5153085 | 0.36 |
ENST00000252321.3
|
KCNA5
|
potassium voltage-gated channel, shaker-related subfamily, member 5 |
| chrX_+_8433376 | 0.36 |
ENST00000440654.2
ENST00000381029.4 |
VCX3B
|
variable charge, X-linked 3B |
| chr18_-_48346298 | 0.35 |
ENST00000398439.3
|
MRO
|
maestro |
| chrY_+_16168097 | 0.34 |
ENST00000250823.4
|
VCY1B
|
variable charge, Y-linked 1B |
| chrX_-_8139308 | 0.34 |
ENST00000317103.4
|
VCX2
|
variable charge, X-linked 2 |
| chrY_-_16098393 | 0.33 |
ENST00000250825.4
|
VCY
|
variable charge, Y-linked |
| chr18_-_48346415 | 0.30 |
ENST00000431965.2
ENST00000436348.2 |
MRO
|
maestro |
| chr3_+_169490834 | 0.29 |
ENST00000392733.1
|
MYNN
|
myoneurin |
| chr16_+_56672571 | 0.25 |
ENST00000290705.8
|
MT1A
|
metallothionein 1A |
| chr8_-_72756667 | 0.25 |
ENST00000325509.4
|
MSC
|
musculin |
| chr1_-_145715565 | 0.24 |
ENST00000369288.2
ENST00000369290.1 ENST00000401557.3 |
CD160
|
CD160 molecule |
| chr8_-_22014339 | 0.24 |
ENST00000306317.2
|
LGI3
|
leucine-rich repeat LGI family, member 3 |
| chr18_-_48346130 | 0.22 |
ENST00000592966.1
|
MRO
|
maestro |
| chr1_-_49242553 | 0.22 |
ENST00000371833.3
|
BEND5
|
BEN domain containing 5 |
| chr14_+_22309368 | 0.21 |
ENST00000390433.1
|
TRAV12-1
|
T cell receptor alpha variable 12-1 |
| chr3_-_108476231 | 0.21 |
ENST00000295755.6
|
RETNLB
|
resistin like beta |
| chr11_+_48002076 | 0.21 |
ENST00000418331.2
ENST00000440289.2 |
PTPRJ
|
protein tyrosine phosphatase, receptor type, J |
| chr8_+_24298597 | 0.21 |
ENST00000380789.1
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr8_-_21669826 | 0.20 |
ENST00000517328.1
|
GFRA2
|
GDNF family receptor alpha 2 |
| chr6_+_31638156 | 0.19 |
ENST00000409525.1
|
LY6G5B
|
lymphocyte antigen 6 complex, locus G5B |
| chr7_-_96654133 | 0.19 |
ENST00000486603.2
ENST00000222598.4 |
DLX5
|
distal-less homeobox 5 |
| chr8_+_24298531 | 0.19 |
ENST00000175238.6
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr9_+_71650645 | 0.18 |
ENST00000396366.2
|
FXN
|
frataxin |
| chr19_-_8408139 | 0.18 |
ENST00000330915.3
ENST00000593649.1 ENST00000595639.1 |
KANK3
|
KN motif and ankyrin repeat domains 3 |
| chr22_+_18632666 | 0.18 |
ENST00000215794.7
|
USP18
|
ubiquitin specific peptidase 18 |
| chr2_-_74875432 | 0.16 |
ENST00000536235.1
ENST00000421985.1 |
M1AP
|
meiosis 1 associated protein |
| chr14_-_23623577 | 0.16 |
ENST00000422941.2
ENST00000453702.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr12_-_10251603 | 0.15 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr14_-_24616426 | 0.15 |
ENST00000216802.5
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr2_+_238395803 | 0.14 |
ENST00000264605.3
|
MLPH
|
melanophilin |
| chr6_+_123317116 | 0.14 |
ENST00000275162.5
|
CLVS2
|
clavesin 2 |
| chr14_+_24616588 | 0.14 |
ENST00000324103.6
ENST00000559260.1 |
RNF31
|
ring finger protein 31 |
| chr8_-_49833978 | 0.14 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr13_-_26452679 | 0.14 |
ENST00000596729.1
|
AL138815.2
|
Uncharacterized protein |
| chr3_-_190167571 | 0.14 |
ENST00000354905.2
|
TMEM207
|
transmembrane protein 207 |
| chr8_-_145634731 | 0.14 |
ENST00000349769.3
|
CPSF1
|
cleavage and polyadenylation specific factor 1, 160kDa |
| chr9_+_131683174 | 0.13 |
ENST00000372592.3
ENST00000428610.1 |
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr16_+_25703274 | 0.13 |
ENST00000331351.5
|
HS3ST4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
| chr1_+_1846519 | 0.13 |
ENST00000378604.3
|
CALML6
|
calmodulin-like 6 |
| chr8_-_49834299 | 0.13 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr3_-_118864893 | 0.13 |
ENST00000354673.2
ENST00000425327.2 |
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr16_-_25269134 | 0.13 |
ENST00000328086.7
|
ZKSCAN2
|
zinc finger with KRAB and SCAN domains 2 |
| chr14_-_106552755 | 0.11 |
ENST00000390600.2
|
IGHV3-9
|
immunoglobulin heavy variable 3-9 |
| chr2_+_238395879 | 0.10 |
ENST00000445024.2
ENST00000338530.4 ENST00000409373.1 |
MLPH
|
melanophilin |
| chr19_-_3786354 | 0.10 |
ENST00000395040.2
ENST00000310132.6 |
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr14_-_106668095 | 0.10 |
ENST00000390606.2
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr19_-_3786253 | 0.09 |
ENST00000585778.1
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr14_-_106866934 | 0.09 |
ENST00000390618.2
|
IGHV3-38
|
immunoglobulin heavy variable 3-38 (non-functional) |
| chr1_+_93811438 | 0.09 |
ENST00000370272.4
ENST00000370267.1 |
DR1
|
down-regulator of transcription 1, TBP-binding (negative cofactor 2) |
| chr3_+_169490606 | 0.08 |
ENST00000349841.5
|
MYNN
|
myoneurin |
| chr15_+_65204075 | 0.08 |
ENST00000380230.3
ENST00000357698.3 ENST00000395720.1 ENST00000496660.1 ENST00000319580.8 |
ANKDD1A
|
ankyrin repeat and death domain containing 1A |
| chr14_-_21493884 | 0.08 |
ENST00000556974.1
ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2
|
NDRG family member 2 |
| chr19_+_56187987 | 0.08 |
ENST00000411543.2
|
EPN1
|
epsin 1 |
| chr17_+_38333263 | 0.08 |
ENST00000456989.2
ENST00000543876.1 ENST00000544503.1 ENST00000264644.6 ENST00000538884.1 |
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr3_-_188665428 | 0.08 |
ENST00000444488.1
|
TPRG1-AS1
|
TPRG1 antisense RNA 1 |
| chr1_+_22979474 | 0.08 |
ENST00000509305.1
|
C1QB
|
complement component 1, q subcomponent, B chain |
| chr10_-_87551311 | 0.08 |
ENST00000536331.1
|
GRID1
|
glutamate receptor, ionotropic, delta 1 |
| chrX_-_24045303 | 0.07 |
ENST00000328046.8
|
KLHL15
|
kelch-like family member 15 |
| chr4_+_37892682 | 0.07 |
ENST00000508802.1
ENST00000261439.4 ENST00000402522.1 |
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr17_-_61959202 | 0.07 |
ENST00000449787.2
ENST00000456543.2 ENST00000423893.2 ENST00000332800.7 |
GH2
|
growth hormone 2 |
| chr4_-_69215699 | 0.06 |
ENST00000510746.1
ENST00000344157.4 ENST00000355665.3 |
YTHDC1
|
YTH domain containing 1 |
| chr16_-_67978016 | 0.06 |
ENST00000264005.5
|
LCAT
|
lecithin-cholesterol acyltransferase |
| chr11_-_114271139 | 0.06 |
ENST00000325636.4
|
C11orf71
|
chromosome 11 open reading frame 71 |
| chr2_+_242577027 | 0.06 |
ENST00000402096.1
ENST00000404914.3 |
ATG4B
|
autophagy related 4B, cysteine peptidase |
| chr3_+_169491171 | 0.06 |
ENST00000356716.4
|
MYNN
|
myoneurin |
| chr2_+_242577097 | 0.05 |
ENST00000419606.1
ENST00000474739.2 ENST00000396411.3 ENST00000425239.1 ENST00000400771.3 ENST00000430617.2 |
ATG4B
|
autophagy related 4B, cysteine peptidase |
| chr11_-_506739 | 0.05 |
ENST00000529306.1
ENST00000438658.2 ENST00000527485.1 ENST00000397615.2 ENST00000397614.1 |
RNH1
|
ribonuclease/angiogenin inhibitor 1 |
| chr5_-_173043591 | 0.05 |
ENST00000285908.5
ENST00000480951.1 ENST00000311086.4 |
BOD1
|
biorientation of chromosomes in cell division 1 |
| chr12_+_66582919 | 0.05 |
ENST00000545837.1
ENST00000457197.2 |
IRAK3
|
interleukin-1 receptor-associated kinase 3 |
| chr2_-_231825668 | 0.05 |
ENST00000392039.2
|
GPR55
|
G protein-coupled receptor 55 |
| chr1_-_10856694 | 0.05 |
ENST00000377022.3
ENST00000344008.5 |
CASZ1
|
castor zinc finger 1 |
| chr17_-_61996192 | 0.05 |
ENST00000392824.4
|
CSHL1
|
chorionic somatomammotropin hormone-like 1 |
| chr3_+_118864991 | 0.05 |
ENST00000295622.1
|
C3orf30
|
chromosome 3 open reading frame 30 |
| chr14_-_106692191 | 0.05 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr19_-_14117074 | 0.05 |
ENST00000588885.1
ENST00000254325.4 |
RFX1
|
regulatory factor X, 1 (influences HLA class II expression) |
| chr3_-_119396193 | 0.04 |
ENST00000484810.1
ENST00000497116.1 ENST00000261070.2 |
COX17
|
COX17 cytochrome c oxidase copper chaperone |
| chr22_+_24030321 | 0.04 |
ENST00000401461.1
|
RGL4
|
ral guanine nucleotide dissociation stimulator-like 4 |
| chr17_-_61996136 | 0.04 |
ENST00000342364.4
|
GH1
|
growth hormone 1 |
| chr14_-_21493649 | 0.04 |
ENST00000553442.1
ENST00000555869.1 ENST00000556457.1 ENST00000397844.2 ENST00000554415.1 |
NDRG2
|
NDRG family member 2 |
| chr8_+_91013577 | 0.04 |
ENST00000220764.2
|
DECR1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr10_+_26932068 | 0.04 |
ENST00000544033.1
ENST00000454991.1 |
LINC00202-2
RP13-16H11.7
|
long intergenic non-protein coding RNA 202-2 RP13-16H11.7 |
| chr10_+_115674530 | 0.04 |
ENST00000451472.1
|
AL162407.1
|
CDNA FLJ20147 fis, clone COL07954; HCG1781466; Uncharacterized protein |
| chr12_-_56848426 | 0.04 |
ENST00000257979.4
|
MIP
|
major intrinsic protein of lens fiber |
| chr6_-_52774464 | 0.04 |
ENST00000370968.1
ENST00000211122.3 |
GSTA3
|
glutathione S-transferase alpha 3 |
| chr1_+_22979676 | 0.04 |
ENST00000432749.2
ENST00000314933.6 |
C1QB
|
complement component 1, q subcomponent, B chain |
| chr7_-_4877677 | 0.04 |
ENST00000538469.1
|
RADIL
|
Ras association and DIL domains |
| chr18_+_56530136 | 0.03 |
ENST00000591083.1
|
ZNF532
|
zinc finger protein 532 |
| chr17_-_3794021 | 0.03 |
ENST00000381769.2
|
CAMKK1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha |
| chr1_+_76262552 | 0.03 |
ENST00000263187.3
|
MSH4
|
mutS homolog 4 |
| chr3_+_118865028 | 0.03 |
ENST00000460150.1
|
C3orf30
|
chromosome 3 open reading frame 30 |
| chr18_+_45778672 | 0.03 |
ENST00000600091.1
|
AC091150.1
|
HCG1818186; Uncharacterized protein |
| chr17_-_61996160 | 0.03 |
ENST00000458650.2
ENST00000351388.4 ENST00000323322.5 |
GH1
|
growth hormone 1 |
| chr19_+_57874835 | 0.03 |
ENST00000543226.1
ENST00000596755.1 ENST00000282282.3 ENST00000597658.1 |
TRAPPC2P1
ZNF547
AC003002.4
|
trafficking protein particle complex 2 pseudogene 1 zinc finger protein 547 Uncharacterized protein |
| chr8_+_91013676 | 0.03 |
ENST00000519410.1
ENST00000522161.1 ENST00000517761.1 ENST00000520227.1 |
DECR1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr3_+_13521665 | 0.03 |
ENST00000295757.3
ENST00000402259.1 ENST00000402271.1 ENST00000446613.2 ENST00000404548.1 ENST00000404040.1 |
HDAC11
|
histone deacetylase 11 |
| chr5_-_179498703 | 0.03 |
ENST00000522208.2
|
RNF130
|
ring finger protein 130 |
| chr17_-_6459802 | 0.02 |
ENST00000262483.8
|
PITPNM3
|
PITPNM family member 3 |
| chr19_+_58570605 | 0.02 |
ENST00000359978.6
ENST00000401053.4 ENST00000439855.2 ENST00000313434.5 ENST00000511556.1 ENST00000506786.1 |
ZNF135
|
zinc finger protein 135 |
| chr16_-_66584059 | 0.02 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr17_+_38498594 | 0.02 |
ENST00000394081.3
|
RARA
|
retinoic acid receptor, alpha |
| chrX_+_2609207 | 0.02 |
ENST00000381192.3
|
CD99
|
CD99 molecule |
| chr10_+_695888 | 0.02 |
ENST00000441152.2
|
PRR26
|
proline rich 26 |
| chr1_-_27693349 | 0.01 |
ENST00000374040.3
ENST00000357582.2 ENST00000493901.1 |
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr11_+_114270752 | 0.01 |
ENST00000540163.1
|
RBM7
|
RNA binding motif protein 7 |
| chr2_-_242576864 | 0.01 |
ENST00000407315.1
|
THAP4
|
THAP domain containing 4 |
| chr11_+_114271314 | 0.01 |
ENST00000541475.1
|
RBM7
|
RNA binding motif protein 7 |
| chr12_+_122241928 | 0.01 |
ENST00000604567.1
ENST00000542440.1 |
SETD1B
|
SET domain containing 1B |
| chrX_+_2609317 | 0.01 |
ENST00000381187.3
ENST00000381184.1 |
CD99
|
CD99 molecule |
| chr11_+_114271251 | 0.00 |
ENST00000375490.5
|
RBM7
|
RNA binding motif protein 7 |
| chr4_+_75311019 | 0.00 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chrX_+_2609356 | 0.00 |
ENST00000381180.3
ENST00000449611.1 |
CD99
|
CD99 molecule |
Network of associatons between targets according to the STRING database.
First level regulatory network of AR_NR3C2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.2 | 1.4 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.1 | 0.9 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.8 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.8 | GO:0036018 | cellular response to erythropoietin(GO:0036018) |
| 0.1 | 0.8 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.1 | 0.3 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 1.9 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 0.2 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.4 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.2 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.6 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.2 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.2 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.2 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.8 | GO:0000795 | synaptonemal complex(GO:0000795) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.2 | 0.9 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 1.8 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.1 | 0.4 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.2 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 1.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |